GALEAS Bladder is a simple, sample-to-report urine test to detect the presence of bladder cancer.
GALEAS Bladder leverages ultra-sensitive, targeted next generation sequencing (NGS) chemistry to interrogate somatic mutations in 23 genes associated with bladder cancer in tumor-derived genomic DNA extracted from a urine sample.
It offers a viable alternative to flexible cystoscopy.
One test for ALL stages of bladder cancer
Delivers sensitivity and specificity equivalent to cystoscopy for both Non-Muscle Invasive Bladder Cancer (NMIBC) and Muscle Invasive Bladder Cancer (MIBC).
Automated and scalable workflow
GALEAS Bladder can be run in any NGS capable laboratory and automated and scaled according to throughput.
Sample to answer solution
From home sample collection through to bioinformatics and reporting, the process is optimized and streamlined to maximize efficiencies.
Part of a suite of products for oncology and liquid biopsy
GALEAS Bladder workflow
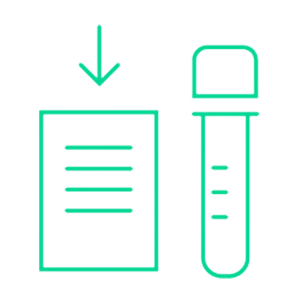
Service laboratory dispatches bar coded GALEAS urine collection device to patient
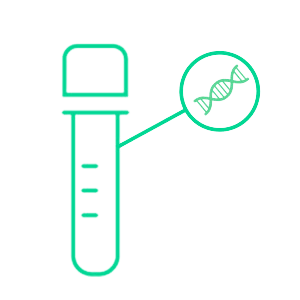
Returned urine sample is centrifuged and gDNA extracted from cell pellet using Bead Xtract urine gDNA extraction kit
NGS libraries are prepared with the GALEAS Bladder kit and sequenced. 5M reads (2 x 150bp PE reads)
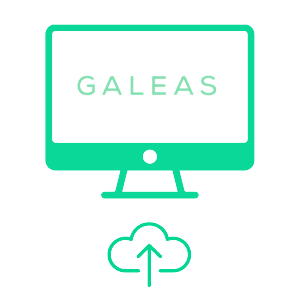
FASTQ files are uploaded to secure cloud-based GALEAS analysis platform
Results are analyzed, report generated and shared directly with requesting physician
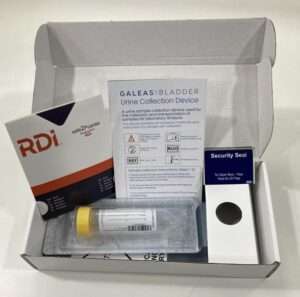
Shipped ready for dispatch to patients, the simple, intuitive cardboard collection device allows patients to provide a urine sample in the comfort of their own home.
The urine sample is returned to the lab in a 50ml Falcon tube containing our proprietary preserver validated with our DNA extraction kit and panels to ensure optimal performance.
The tube is barcoded to provide full traceability.
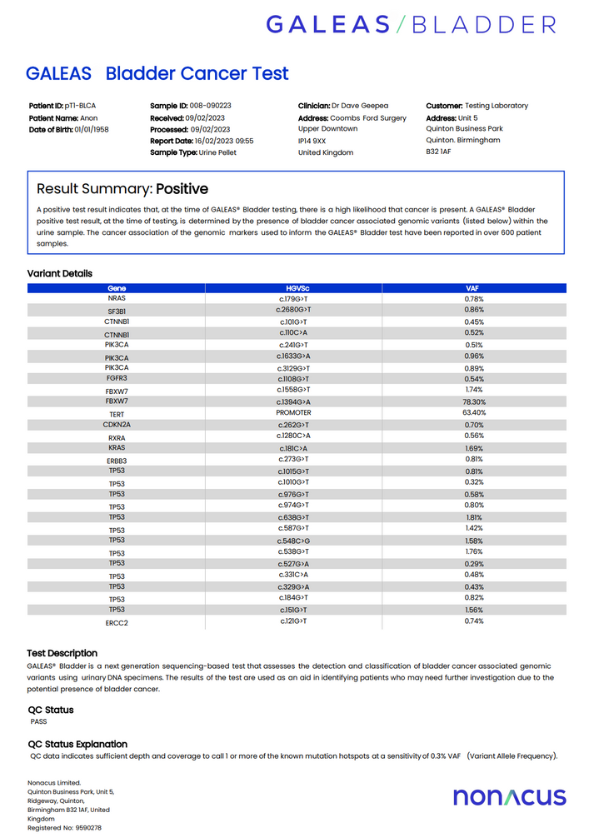
GALEAS Bladder enables the highly sensitive and accurate detection of tumor-derived DNA across all stages and grades of bladder cancer
GALEAS Bladder accurately detects somatic mutations present in over 96% of bladder cancer cases and has been validated using 710 urine samples from three UK clinical cohorts.1
| Sensitivity | Specificity | |
|---|---|---|
| pTa | 89% | 86% |
| T1 | 97% | 86% |
| T2+ | 92% | 86% |
| G1 | 78% | 86% |
| G2 | 93% | 86% |
| G3 | 96% | 86% |
| NMIBC | 92% | 86% |
| MIBC | 92% | 86% |
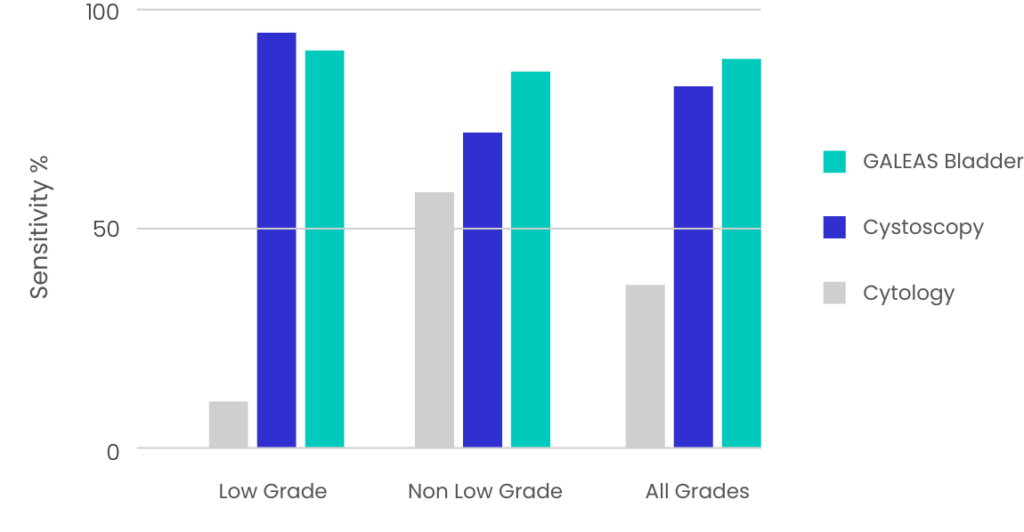
Figure 1: GALEAS Bladder performance across all stages of bladder cancer and compared to reported sensitivity of cytology and cystoscopy.
Why choose GALEAS Bladder?
Simple end to end workflow for easy implementation
From ready to ship urine collection devices through to secure cloud-based reporting the process is optimized to maximize efficiencies.
Optimized reagents ensure reliability of test results
All reagents associated with the GALEAS Bladder workflow have been developed and optimized to ensure consistent and reliable results.
Flexible throughput offers scalability
By covering a wide range of markers, GALEAS Bladder can be used for patient monitoring, relapse, MRD surveillance and companion diagnostics as well as hematuria triage.
GALEAS Bladder is for research use only and not for use in diagnostic procedures outside of the UK. Our UKCA marked product can be found here: GALEAS Bladder (UKCA)| Nonacus
References
1. Ward DG, Baxter L, Ott S, Gordon NS, Wang J, Patel P, et al. Highly sensitive and specific detection of bladder cancer via targeted ultra-deep sequencing of urinary DNA. European Urology Oncology. 2023 Feb;6(1):67-75.