The power of SPRI technology: Tips for DNA size selection and effective clean-up in NGS workflows
By Victoria Simms on November 1, 2023 | Reviewed by Celina Whalley
Since its introduction into genomics in 1995, Solid Phase Reversible Immobilization (SPRI) technology, in the form of magnetic beads, has become mainstream in laboratory protocols for nucleic acid extraction and purification.
SPRI beads are highly abundant, cost-efficient and have time-saving capabilities by being automation-friendly.1
This blog will explore:
- The key features and powerful benefits of SPRI technology
- SPRI bead types and how they are used in NGS workflows
- Tips to help you generate high-quality samples when using magnetic beads for:
- DNA cleanup steps
- DNA fragment sizing
An introduction to SPRI technology
The technology uses paramagnetic particles, often known as beads, to bind nucleic acids reversibly and selectively. By modifying the magnetic bead properties, it is possible for nucleic acids to be extracted, purified and size selected accurately and reproducibly.
SPRI technology produces high-quality samples that are suitable for a wide range of downstream applications including: quantitative PCR (qPCR), droplet digital PCR (ddPCR), microarray analysis, molecular and immunodiagnostics, magnetically activated cell sorting (MACS), Sanger and Next Generation Sequencing (NGS).
Bead applications within genomics are wide-ranging and depending on the workflow you are using, you may come across multiple types in one protocol.
The key features of SPRI beads
Most SPRI beads are around 1 µm in size, which provides a large solid surface area for nucleic acid binding. The bead core usually consists of light-weight polystyrene, coated with magnetite (see Figure 1). This is responsible for giving the bead its paramagnetic property – meaning it is only magnetic within a magnetic field.
This feature is utilized within SPRI technology protocols, such as NGS cleanups, to easily separate the DNA bound to the bead surface from other contaminants and unwanted material from within the sample and ensures that the beads do not clump together in solution.
There are different types of SPRI beads and the chemistry of the outer layer of the bead is adapted depending on the intended purpose of the bead, ie, for PCR purification, NGS cleanup or DNA size selection. However, their main function is to facilitate nucleic acid binding by providing a solid surface and access to carboxyl groups.
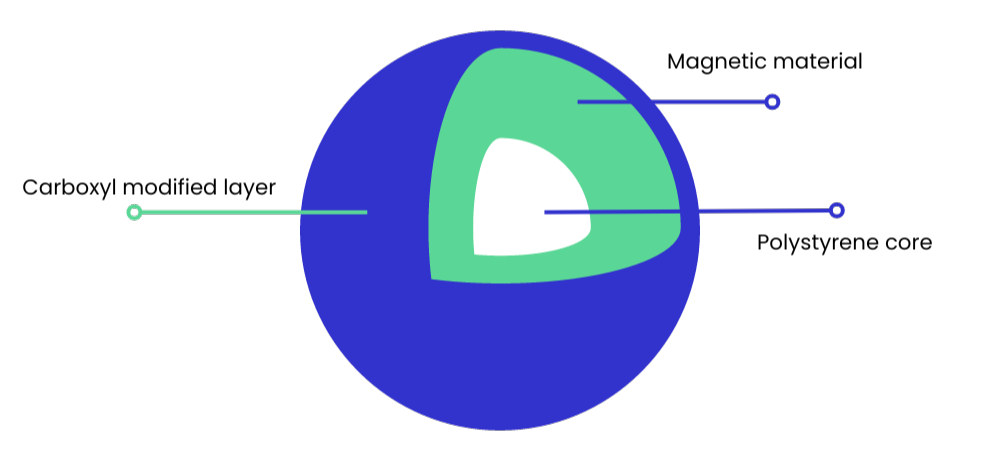
Figure 1: Typical structure of a carboxyl magnetic SPRI bead.
The benefits of SPRI beads
- Paramagnetic particles – they are magnetic only within a magnetic field; meaning that the beads and bound material can be easily separated from any contaminants and that the beads are not attracted to each other in solution.
- No requirement for filtration or centrifugation - which can result in loss of sample.
- Compatible with automation equipment - reducing hands-on time.
- Relatively uniform bead size of 1 µm – reproducible results and minimal particle settling in solution. This feature is particularly important for automation.
- Lightweight core – remains suspended in solution.
- Fast magnetic response – rapid nucleic acid purification.
- Negatively charged solid outer surface – the carboxyl groups present on the solid support quickly and effectively bind nucleic acid fragments.
- High surface area to mass ratio – the small bead size results in high surface area to mass ratio, which achieves high binding capacity for the nucleic acid fragments, which is important for high yield.
Types of SPRI beads
NGS library preparation protocols use SPRI technology as magnetic bead separation can be performed relatively easily. Other than access to a magnet, no specialized laboratory equipment is needed and there is no requirement for centrifugation or filtration.
So, what are the differences between the various types of SPRI beads? And what do you need to consider when working with each of them?
See Table 1 for a list of comparisons between clean-up and size selection beads and read our tips below on how to optimize the bead performance to enhance the quality of your sequencing results, when working with SPRI technology.
Library preparation protocols often use three different types of beads:
- PCR/NGS cleanup beads ie, AMPure XP
- Nucleic acid size selection ie, SPRIselect
- Library capture beads ie, Streptavidin-coated beads ie, dynabeads
Table 1: Comparisons between the SPRI beads involved in PCR/NGS clean-up and DNA size selection.
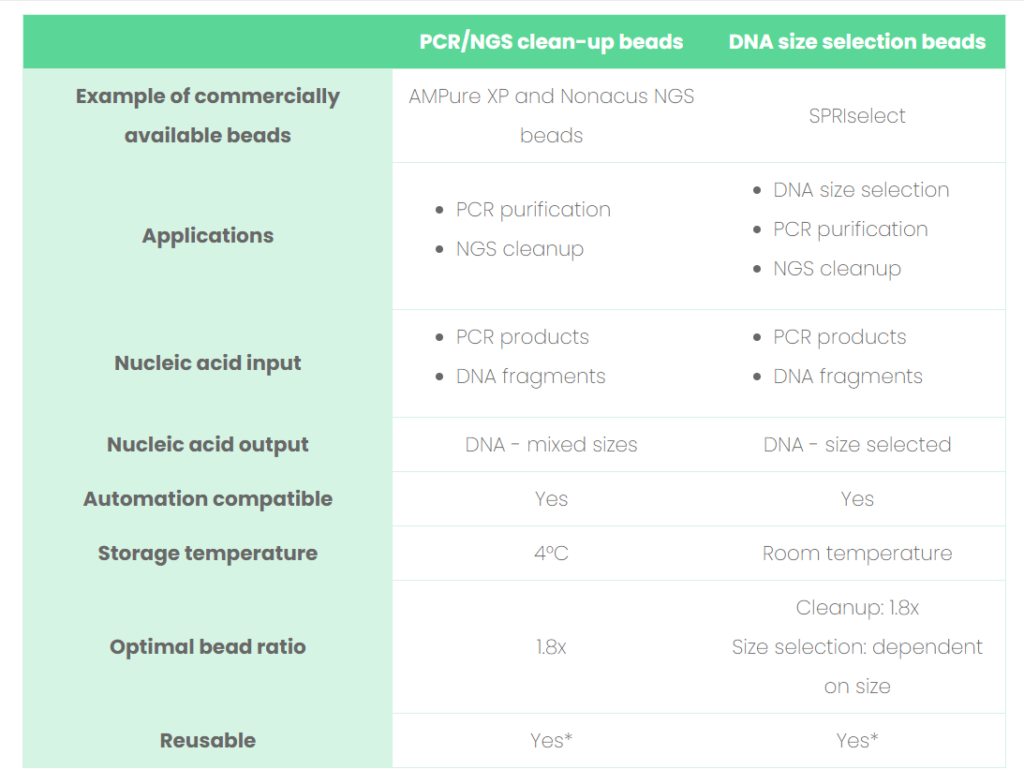
*Would advise only when cross-contamination is not a concern. Always refer to the bead manufacturer’s instructions.
Tips for SPRI bead clean-up
Read on, and then watch our video to see bead clean-up in action.
Before beginning a clean-up protocol (see Figure 2), it may be necessary to buy, aliquot and wash the beads - always refer to the manufacturer’s guidance before starting the workflow.
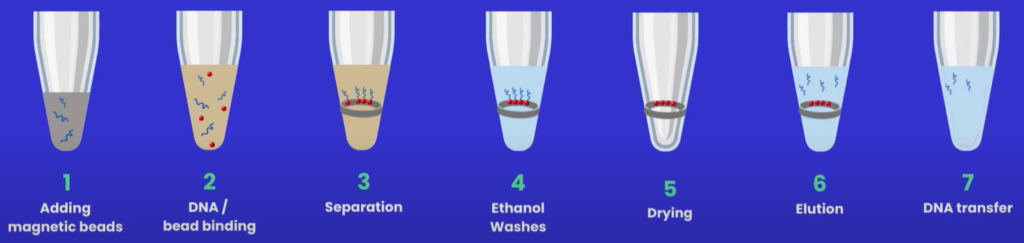
Figure 2: Magnetic bead clean-up protocol.
Pre-preparation tip: Ensure the beads have been stored at the correct temperature before beginning. They then need to be fully resuspended in solution to ensure the correct amount of beads is used in the protocol.
The paramagnetic beads are added to your sample of interest for nucleic acid extraction.
Tip: After adding the SPRI beads to your sample. Be sure to mix well so that the solution is homogenous, to ensure efficient DNA binding. Insufficient mixing at this stage will decrease your final yield.
Protocol Step 1: Adding magnetic beads
Protocol Step 2: DNA binds to the magnetic beads
The surface of the magnetic bead has been designed and optimized to ensure efficient binding of the target fragments. The high surface area and carboxyl groups on the bead surface make bead-nucleic acid interactions possible.
Protocol Step 3: Separation
An external magnetic field is introduced, which attracts the magnetic beads and bound target material. The beads move to the side of the tube where they are immobilized until the magnetic field is moved away.
Tip: Ensure the correct magnet is used for the tubes or plates that you are using. Be aware of the differences between skirted, semi-skirted and non-skirted plates as they will require specifically designed magnets to accommodate the skirting as the beads to be fully immobilized to achieve a high DNA yield.
Protocol Step 4: Ethanol washes
Any unwanted components ie, proteins, other nucleic acids, cellular and tissue components are removed using a series of washing steps. During these washes, the beads remain immobilized to side of the tube, whilst the supernatant is discarded.
Tip: Make up the ethanol solution up on the day of performing the protocol, ethanol will evaporate causing the concentration to change.
Tip: Use a volumetric cylinder or serological pipette to measure out the amount of ethanol required for the washing steps. The density difference between ethanol and water can lead to inaccurate measurements when making up percentages of ethanol ie, 80% ethanol.
Protocol Step 5: Drying
Ethanol is removed from the beads, using a pipette and the beads are left to air dry.
Tip: Ensure all excess ethanol is removed, this can be achieved by doing a quick spin during the bead drying time and then pipetting off the excess ethanol. Any residual ethanol in the final sample will affect the NGS performance.
Tip: Monitor the appearance and condition of your beads throughout the drying time to know when to elute. You want to elute when the beads are matte in appearance (Figure 3). If cracks appear, still proceed but ensure you resuspend the beads fully by vigorous pipetting.
Tip: note your surrounding temperature and whether there is air conditioning in the room you are in – these factors will affect the drying time. Elute based on the appearance of the beads and not necessarily the exact time stated in the protocol – the time stated is there as a guide.
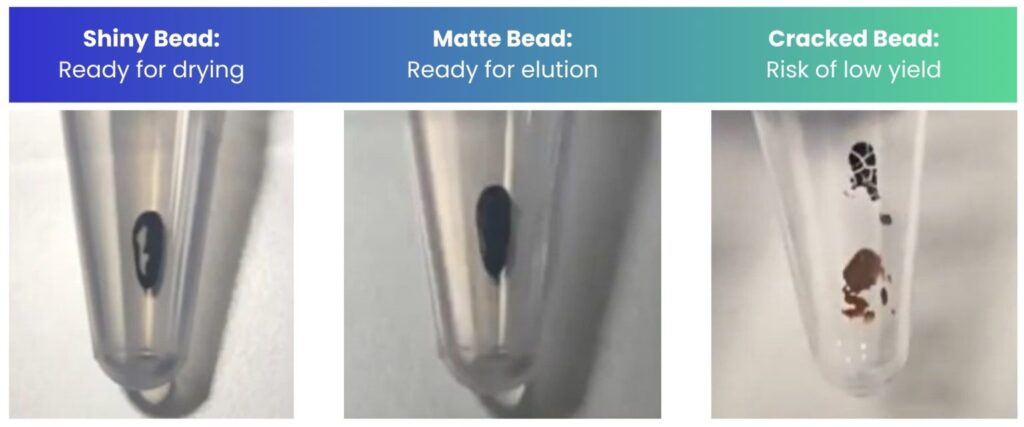
Figure 3: The appearance of the beads during the drying step of bead clean-up.
Protocol Step 6: Elution
Elution buffer is added to the bead/DNA complex and the buffer properties release the DNA from the beads into solution. A sufficient volume of buffer is required to fully elute your sample; however care must be taken to not use excess volumes as this will dilute your sample, which may cause challenges in downstream applications.
Tip: If you are processing/eluting many samples at once, it may be necessary to add elution buffer to your all of your samples to prevent them from drying out, before resuspending and then placing them back onto the magnet.
Protocol Step 7: DNA transfer
The magnetic field pulls the beads away leaving the purified DNA in the buffer, which can then be removed into a new tube. The DNA sample has now been purified and is ready for subsequent quality control and downstream analysis as guided in your NGS protocol.
Tips for SPRI bead DNA size selection
SPRIselect beads are often used to produce a uniform distribution of double-stranded DNA fragments during NGS library preparation, which is critical for quick and reliable downstream sequencing processing and analysis.
This technique is best applied to fragments between 150-800 bp and can be easily implemented using automation. In this way, the need to size select via chips or gels is eliminated.
The workflow for DNA size selection will be dependent on the fragment size (in bp) that you wish to capture.2
- Single-sided - with an increase in bead:sample ratio comes an increase in the efficiency of binding smaller fragments giving a broader range of DNA fragments in the final elution solution.
- Double-sided has an additional capture step - large fragments are removed in the first step, and smaller fragments in the second, so there is a variation of bead:sample ratio depending on the final library size required.
Below is a schematic overview of the DNA sizing protocols (Figure 4) which highlights the differences, along with associated tips.
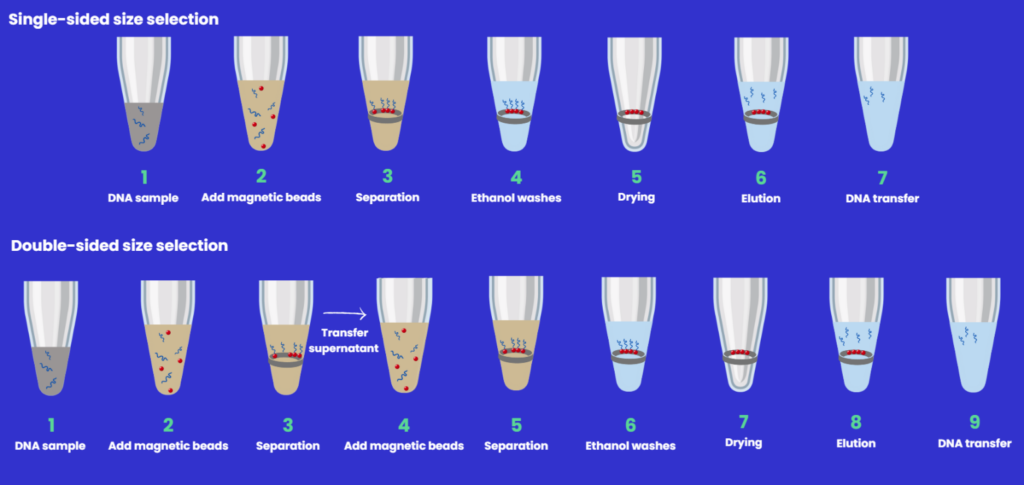
Figure 4: Single and double- and right-sided size selection workflows.
Tip: Before starting.
Particular attention should be given to the SPRI beads before starting the protocol to make sure they are fully resuspended.
Tip: Binding step.
When adding the SPRIselect beads to your sample - make sure to fully mix the combined solution so that the mixture is homogenous. Insufficient mixing will lead to inconsistent size selection.
Tip: Separation steps.
Allow the SPRI beads to fully settle on the magnet. A large sample volume, a high ratio of SPRI beads or weak magnet will require a longer time for the beads to be pulled to the magnet. Making sure the beads are fully immobilized will ensure that you can remove the supernatant with minimal bead transfer. Losing beads and bound material at this stage, would decrease your final yield.
Tip: DNA elution step.
Ensure the beads are fully covered with elution buffer and that the beads are fully immobilized before removing elution buffer, as any beads carried over impacts the the final sample purity.
Commercially available SPRI beads have increased in availability and continue to do so. They are often included in NGS library preparation kits, or you can buy in your own stock from an external supplier. As a result, you can fall into the idea they have specific roles, when they just differ in their trade names.
Working with SRPI-SPRI beads – are they interchangeable?
Both NGS clean up and DNA size selection beads can be used for PCR purification as well as NGS clean-up (see Table 1). However, the storage conditions of these two types of beads are different and the intended use of these two bead types vary; as such, these bead types have been designed and validated differently.3
- Size selection beads (eg, SPRIselect) are guaranteed to result in a particular DNA size in relation to input volume. This allows accurate and consistent DNA size selection from lot-to-lot, but this comes at a price, so it’s not cost effective to use these for both DNA size selection and clean-up steps routinely
- DNA cleanup beads (eg, AmpureXP) are not validated to the same degree, as size-selection beads, as their intended use is to capture a broad DNA size range – yes, they can be used for size selection too, but it would be wise to test each lot at different bead ratios against a ladder to ensure reproducibility (substituting the clean-up beads 1:1 with size selection beads)
- Note that streptavidin beads (eg Dynabeads M-270 Streptavidin beads) are quite different. They are designed to take advantage of the attraction between streptavidin and biotin. and are not interchangeable and should only be used for DNA capture where specified in the NGS workflow. This is because their intended use is to isolate biotinylated nucleic acids.
SPRI technology at Nonacus
This blog has provided an insight to SPRI bead technology and an overview, with tips, of its use in relation to DNA cleanup and size selection applicable to NGS library preparation.
Here at Nonacus we specialize in NGS library preparation incorporating SPRI technology into our Cell3 Target™ technology, with a focus on cell-free DNA from liquid biopsy.
References
-
- Deangelis MM, Wang DG, Hawkins TL. Solid-Phase Reversible Immobilization for the Isolation of PCR Products. Vol 23.; 1995.
- SPRIselect Beads | PCR & NGS Size Selection and Cleanup. Accessed October 12, 2023.
- SPRIselect vs AMPure XP beads—What is the difference? Accessed October 12, 2023.