Focused exonic content
Optimized exome coverage provides 33 Mb of highly-conserved protein coding regions and 99% of ClinVar variants, so you can focus on what matters.
Supports a range of sample input types
Accommodates a broad range of sample types; cell-free (cfDNA), cell-free fetal (cffDNA), and DNA from fresh frozen as well as formalin fixed paraffin embedded (FFPE) tissue.
Optimized performance
Delivering excellent uniformity of coverage, minimizing low coverage regions, thereby reducing sequencing costs and improving sample throughput.
Customizable content
Designed to be flexible, Cell3 Target: Whole Exome allows you to add extra content to cover targets specific to your requirements.
Whole exome enrichment
Whole exome sequencing has been widely adopted in the last decade as an efficient way of screening the genome for disease-associated mutations.
By focusing reads on coding regions, which contain more than 80% of known disease-causing variants, the probability of identifying mutations associated with disease is increased. At the same time, the amount of exome sequencing, and analysis required, is reduced by 99% when compared to sequencing whole genomes, significantly reducing the cost of sequencing.
This makes whole exome sequencing an efficient and cost-effective alternative to whole-genome sequencing especially in clinical applications.
Exome content focused on what matters
The Cell3 Target: Whole Exome focuses on the core protein-coding regions referenced in CCDS. Using a 33 Mb design (37 Mb sequencing footprint) covering 99% of ClinVar variants, we achieve coverage of >97% of targeted regions at >= 20× coverage with a 150x mean sequencing depth and requiring just 4.90 Gb of sequencing per sample.
By focusing on what matters, the Cell3 Target: Whole Exome offers you a cost effective and efficient whole exome solution and the choice of running more samples or samples at greater depth, than other exome products.
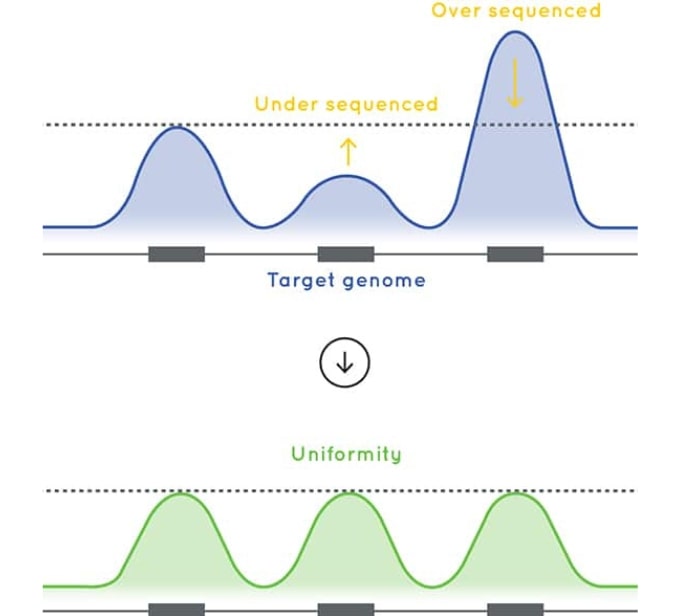
Optimized exome sequencing performance
The baits used in Cell3 Target: Whole Exome are designed to deliver excellent uniformity of coverage. By improving uniformity of coverage and reducing the number of low-coverage exons, our whole exome enrichment optimizes sequencing efficiency and sample capacity per sequencing run.
Supports a range of sample input types, including FFPE DNA
You may want to use the same exome product on multiple sample types – especially for testing matched samples. The Cell3 Target: Whole Exome sequencing kit has been developed for a broad range of DNA input types covering; cfDNA, cffDNA, DNA from fresh frozen and FFPE tissue.
Using as little as 1 ng of genomic or cfDNA (10 ng of FFPE), Cell3 Target: Whole Exome unlocks the door for prenatal and oncology applications.

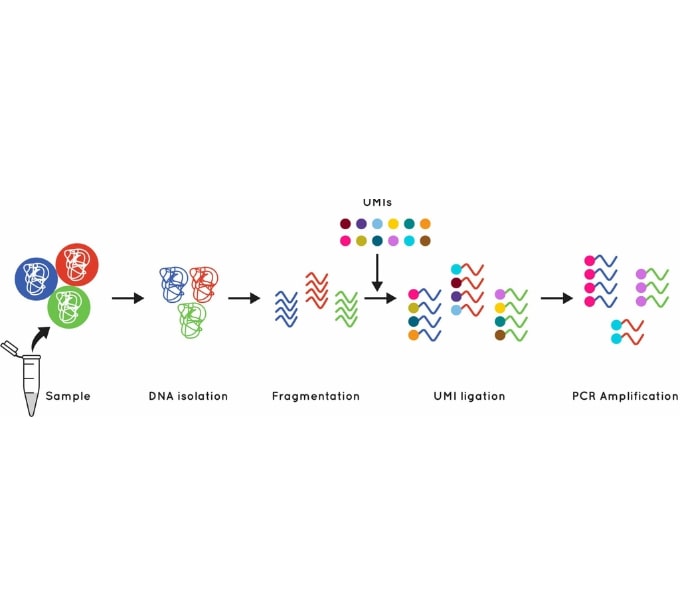
Figure 1: Using UMI’s to identify and quantify individual DNA molecules during library preparation increases sensitivity.
Detect low frequency mutations for prenatal or cancer research
The Cell3 Target library preparation behind our whole exome enrichment incorporates error suppression technology. This includes unique molecular indexes (UMIs) and unique dual indexes (UDIs), to remove both PCR and sequencing errors and index hopping events.
This error suppression technique, combined with our excellent uniformity of coverage, allows you to confidently and accurately call mutations down to 0.1% VAF and enables generation of exome sequencing libraries from as little as 1 ng cfDNA input.
Customizable content on whole exome products
Designed to be flexible, our Cell3 Target: Whole Exome allows you to add extra content specific to your project. Whether this is additional content or increased coverage of existing content, our Panel Design Tool makes this a simple and easy process to implement. And our rapid production turnaround means you will receive a fully NGS-validated custom exome panel within 4 weeks.
- Additional content with high enrichment uniformity
- Increased coverage of specific genes covered by Cell3 Target: Whole Exome
- Optimization of spike-in ratio
Start your custom design now using the Panel Design Tool
For Research Use Only. Not for use in diagnostic procedures.
See our customer publications
Cell3™ Target panels are available with one of two versions of our library preparation kits:
- Fragmentation: for use with DNA (genomic, FF, FFPE)
- Non-fragmentation: for use with cell-free DNA
| Product | Catalog No. |
| Cell3™ Target: Whole Exome, Frag 16 samples | NGS_C3T_WEX_FR_16 |
| Cell3™ Target, Whole Exome, Frag 96 samples | NGS_C3T_WEX_FR_96_A/B/C/D* |
| Cell3™ Target, Whole Exome, Non Frag 16 samples | NGS_C3T_WEX_NF_16 |
| Cell3™ Target, Whole Exome, Non Frag 96 samples | NGS_C3T_WEX_NF_96_A/B/C/D* |
* To provide flexibility in multiplexing samples, our 96-sample kits offer a choice in adapter plate:
A = Adapter plate with indexes 1-96
B = Adapter plate with indexes 97-192
C = Adapter plate with indexes 193-288
D = Adapter plate with indexes 289-384