Comprehensive genomic profiling of clinically relevant cancer biomarkers
GALEASTM Tumor (UKCA)
Enhanced comprehensive genomic profiling (CGP) of clinically relevant biomarkers at your fingertips
Expertly curated content delivered through a single workflow for CGP
GALEAS Tumor is an expertly curated NGS panel developed in parallel with bioinformatics pipelines for the comprehensive profiling of solid tumors. It targets 519 carefully selected, clinically relevant genes aligned with the UK NHS national genomic test directory, NCCN, FDA and ESMO guidelines.
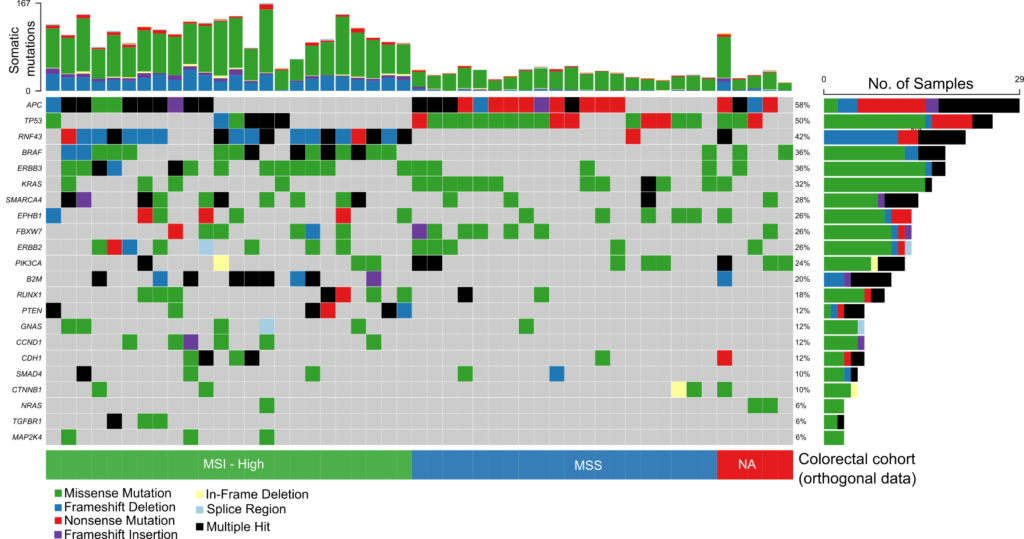
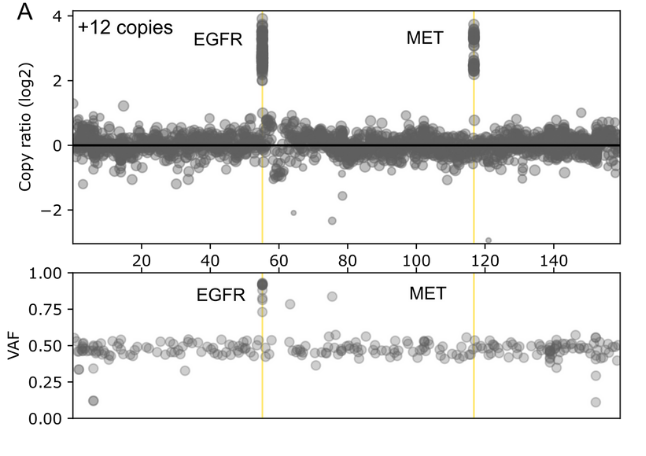
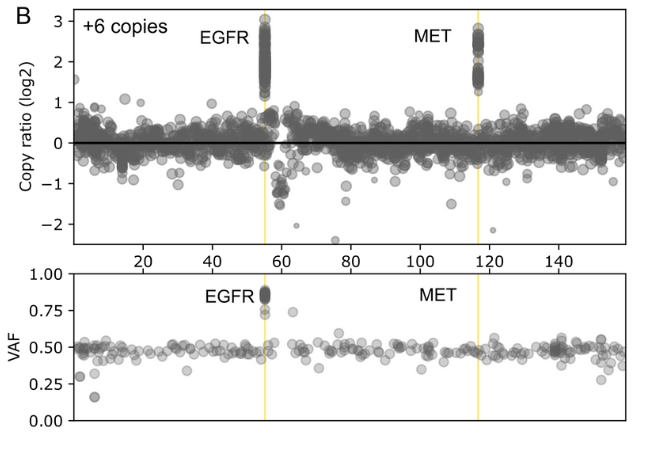
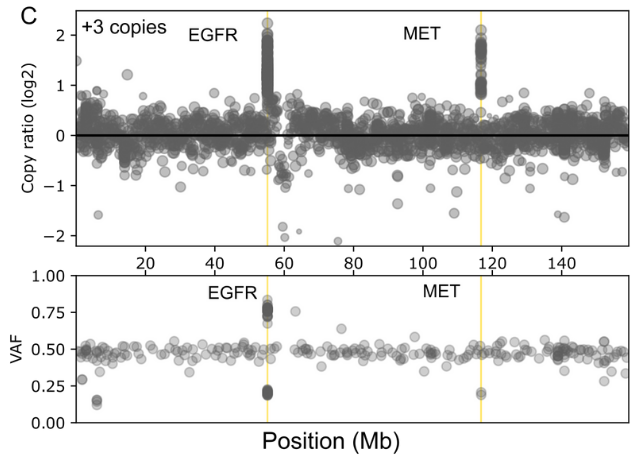
The comprehensive design supports stratification of both common and rare cancers in a single workflow and enables confident detection of a wide range of gene aberrations known to drive cancer including SNVs, INDELs, selected fusions and genome-wide CNVs.
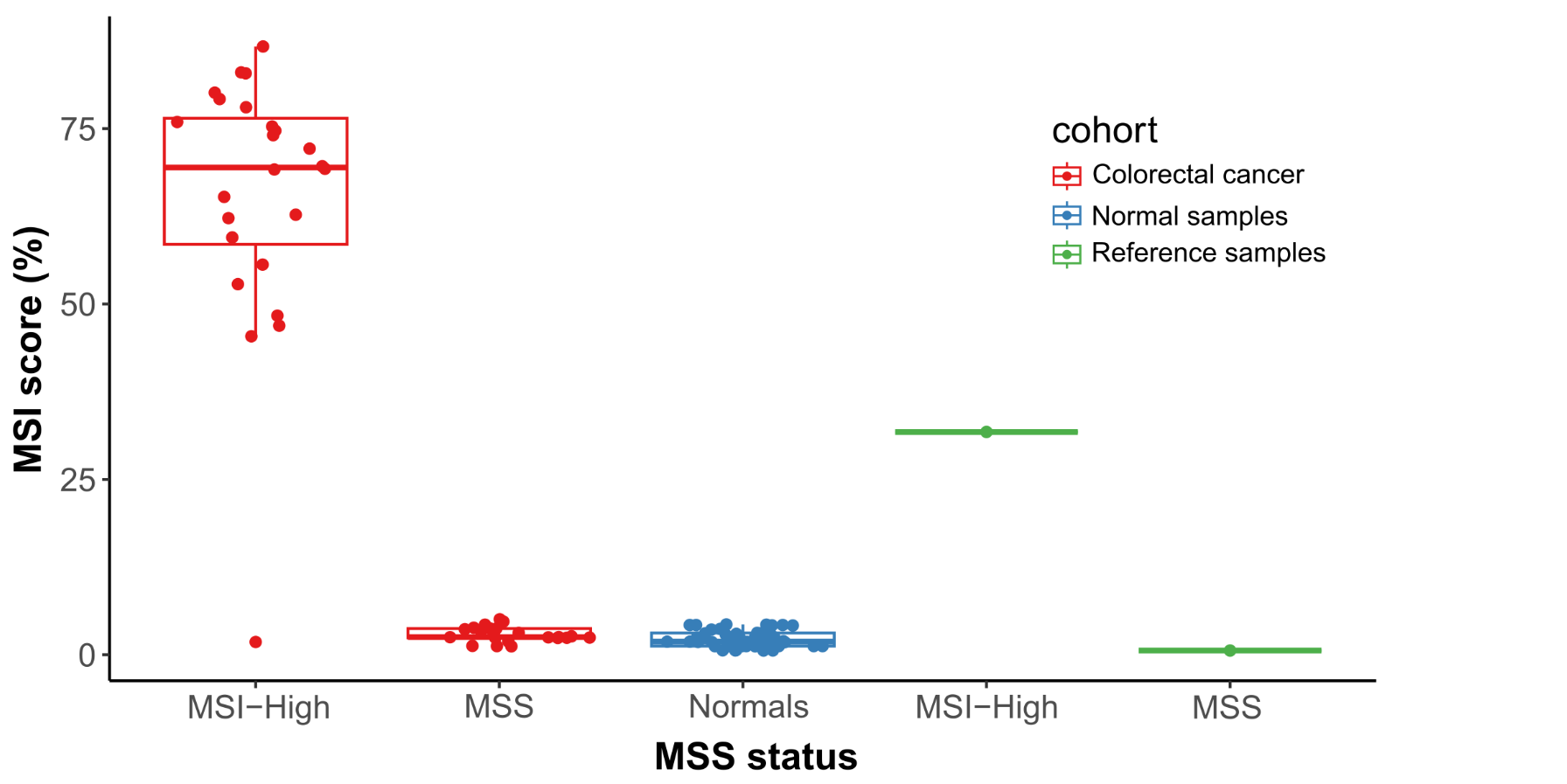
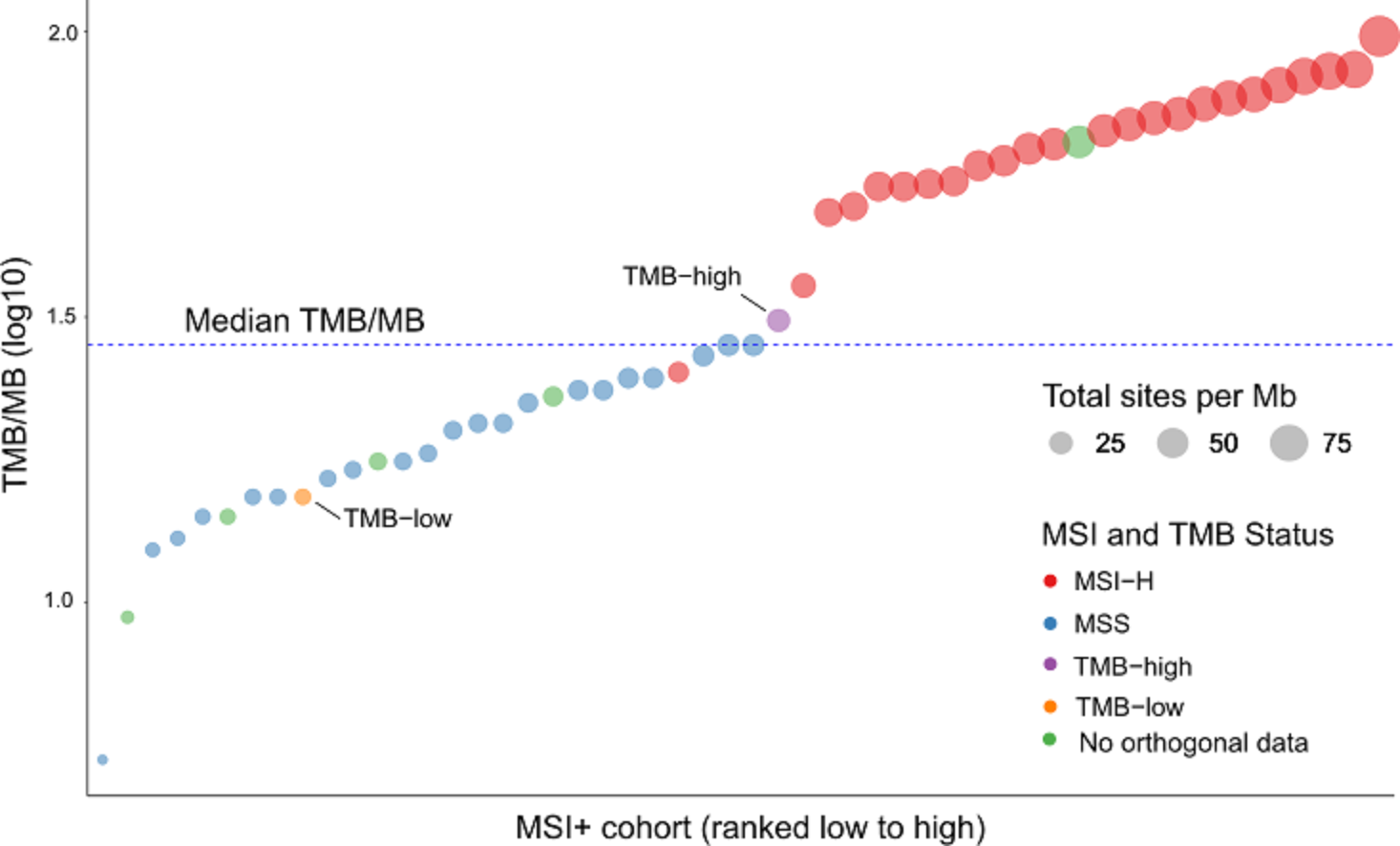
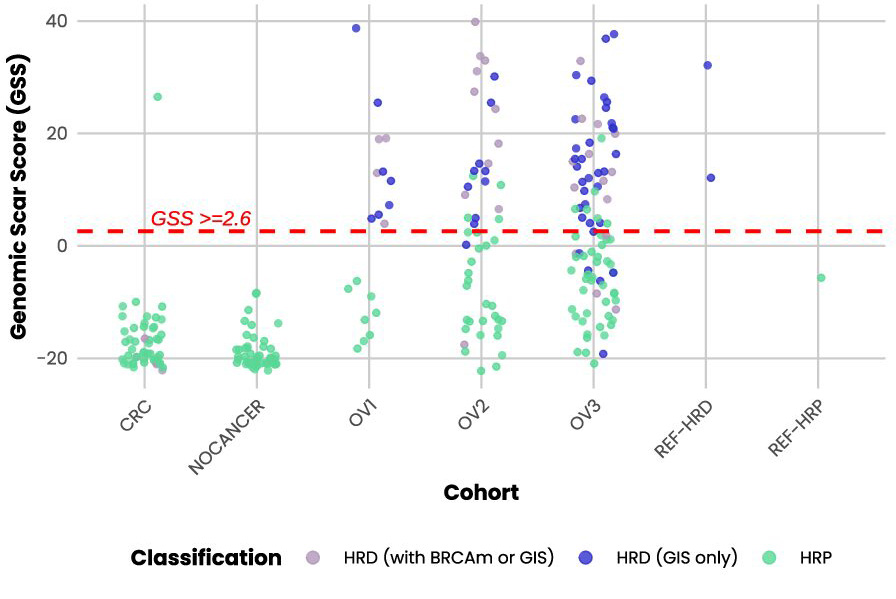
Probe enhancements support the assessment of other biomarkers like Tumor Mutational Burden (TMB), Microsatellite Instability (MSI) and Homologous Recombination Deficiency (HRD).
Key Features
- Targets 519 genes covering rare and common cancers including hereditary and pediatric cancers.
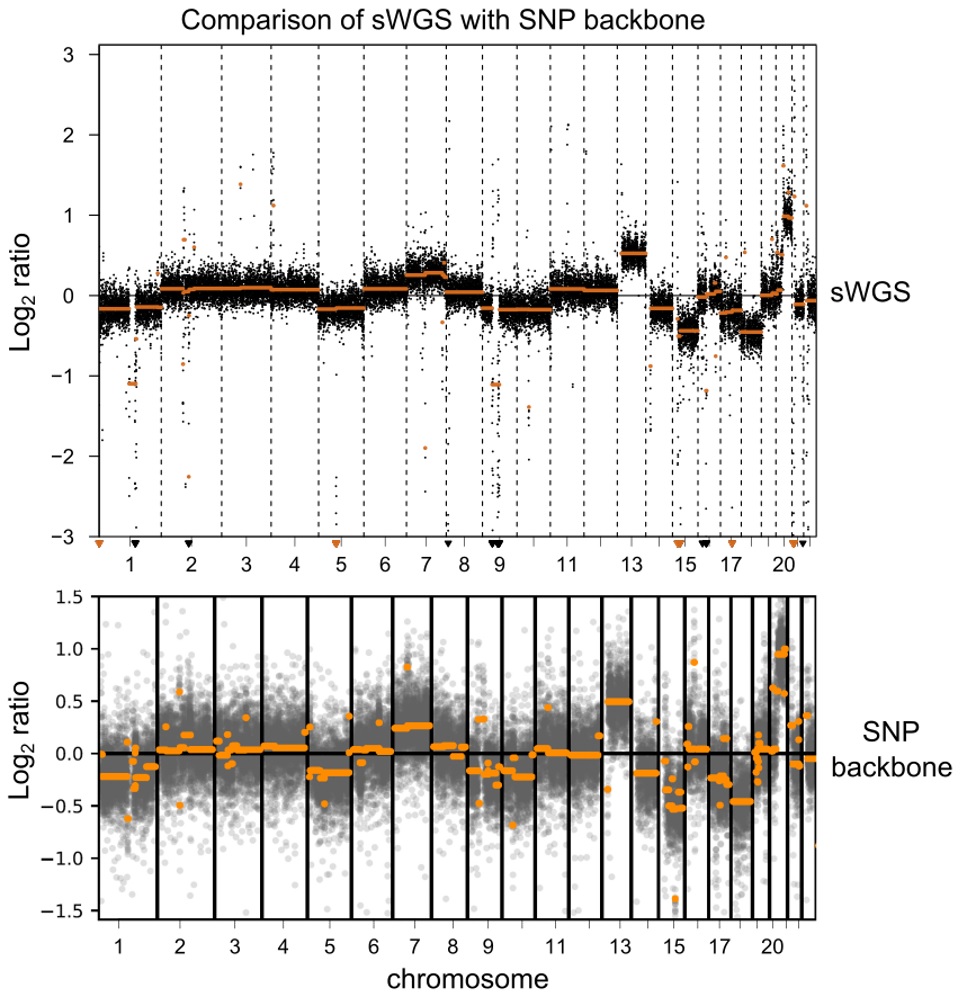
- Genome-wide SNP backbone delivers robust and reliable CNV calling.
- Probes enhancements allow MSI and TMB Immuno-oncology biomarker scoring.
- BRCA variant calling and GI scoring deliver HRD assessment in a single workflow.
- Enhanced coverage of the 1p/19q co-deletion associated with Glioma supports brain cancer studies.
- Coverage of 10 Fusion/Structural rearrangements: ALK, BRAF, EGFR, FGFR2, FGFR3, NTRK1, NTRK2, RET, ROS1, TMPRSS2 reduces need for additional workflows.
- Sample identity tracking SNPs reduce potential errors in laboratory processes.
- Target 64 Pharmacogenomics (oncology) markers.
- Incorporates HLA design relevant for solid tumors.
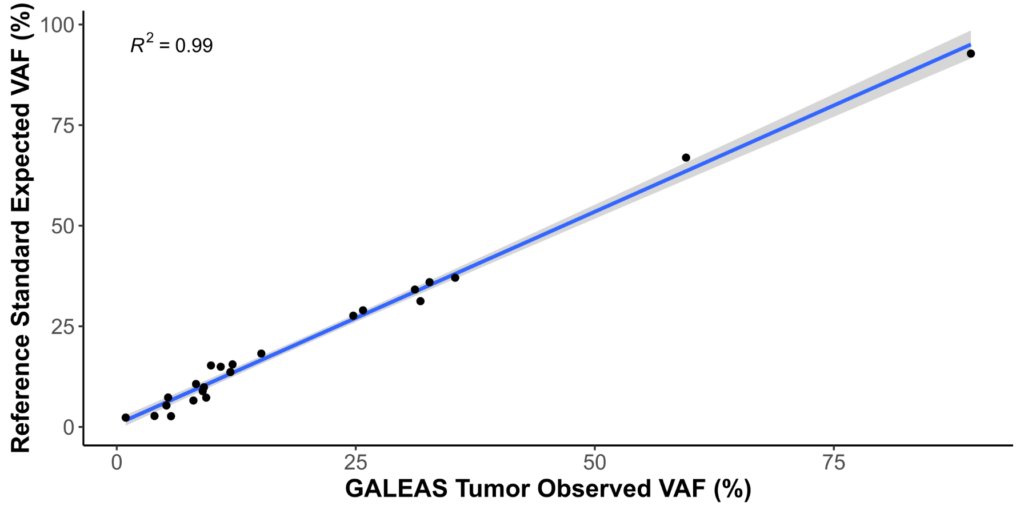
Performance
Why choose GALEAS Tumor?
Current and clinically relevant content
Detect SNVs, INDELs, selected fusions, CNVs, TMB, MSI and HRD associated with a wide range of cancers from common to rare in a single workflow.
Highly efficient capture chemistry
High on-target rates and excellent uniformity of coverage deliver more efficient sequencing and better sensitivity for comprehensive genomic profiling.
Supported by GALEAS software
Developed in parallel with the GALEAS Tumor panel, our GALEAS analysis software provides users with optimised bioinformatics for accurate variant calling and supports rapid integration into routine use.
GALEAS Tumor is UKCA marked in accordance with the UK Medical Devices Regulations 2002 (as amended). Nonacus Ltd. is the legal manufacturer and is responsible for the development and ascertainment of the performance characteristics of GALEAS Tumor.
The GALEAS Tumor workflow is simple and easy

gDNA, FFPE DNA samples
Wide range of sample types including FFPE, frozen tissue and blood

Sample preparation
DNA extraction kits

Prepare libraries and enrich
GALEAS Tumor

Sequence
Illumina NGS Sequencing System

Call variants
GALEAS analysis software

Report
Generate reports for HRD, TMB and MSI

Interpret and report
Utilize third party tertiary software for interpretation and reporting
Optimised Library Preparation
GALEAS Tumor library preparation is simple and easy compared to other targeted sequencing methods for hybridization and capture.
It requires as little as 10 ng of DNA and takes less than 10 hours, with less than two hours hands-on time. It is designed with multiple stop points to provide flexibility within laboratory processing. Library preparation can be run manually or automated (up to 96 samples in a single batch). Indexes are available for up to 384 samples to facilitate high throughput laboratories and to allow for flexible batch sizes.
Turnkey bioinformatics
Complimentary access to GALEAS cloud-based software delivers high quality variant calling and genome wide scores
Cutting-edge bioinformatics pipelines were developed in tandem with, and specifically for GALEAS Tumor ensuring superior detection of all key variant types (SNV, INDEL, CNV and SV’s) as well as genome wide scores for MSI and TMB and assessment of HRD. This turn-key software solution requires no specialist bioinformatic knowledge, enabling labs to rapidly integrate GALEAS Tumor into routine use.
Including pre-validated, optimised variant calling software with GALEAS Tumor gives clinical scientists confidence that they can deliver the robust, reliable and clinically actionable information that is critical for downstream interpretation.
Technical performance
High on-target rates and excellent uniformity of coverage deliver more efficient sequencing for comprehensive genomic profiling
GALEAS Tumor uses the Cell3 Target library preparation technology optimised by Nonacus to deliver a high percentage of on-target reads, low duplication rates and more uniform coverage.
This exceptional technical performance means lower DNA inputs, less sequencing and higher recall and precision across more variants.
| Key quality indicator | GALEAS Tumor |
|---|---|
| Number of genes | 519 |
| Capture Panel Size (Mb) | 3.74 Mb |
| Gb required for mean 500x coverage (2x100bp PE) | 5 Gb |
| Percentage coverage ≥250x | 98% |
| Percentage on or near bait | 71% |
| Percent duplication | 9% |
| SNV recall | 100% |
| INDEL recall | 100% |