FFPE, cfDNA and gDNA: The guide to DNA sample types used in next generation sequencing
By Celina Whalley on December 7, 2023. Reviewed by Victoria Simms.
Next-generation sequencing (NGS) has had a game-changing impact on genetic research since its introduction in 2000, moving the technology forward to become the primary platform used for de-mystifying the genetic code.
It is defined as the sequencing of small DNA fragments in a massively parallel fashion and, as it enables the analysis of many different genetic variants in a single assay, it has been accepted as a clinical tool across multiple disciplines.1
To help you achieve optimal sequencing results, this blog will cover:
- Methods to assess the quantitative and qualitative characteristics of the three types of DNA used in NGS workflows:
- Overview of qualitative characteristics – typical electropherogram profiles for each DNA type.
- How DNA quantitative and qualitative characteristics influence your choice of commercially available NGS assay.
The importance of DNA type selection
In recent years, NGS has evolved to become more sensitive and gene-targeted, leading to a reduction in costs alongside the number of tests and surgical procedures needed for diagnosis and prognosis. It has also become an aid to the treatment decision-making process. However, the sequencing assay of choice needs to be made with the type of DNA in mind that’s been extracted to ensure successful NGS analysis.
In alignment with NGS development, commercially available DNA extraction kits now accommodate a wide range of starting material providing clinicians and researchers with multiple options to diagnose, monitor and study disease. For instance, obtaining a urinal sample is faster and less painful for the patient than taking a bladder tissue biopsy, and genetic testing is now so sensitive bladder cancer genes can be detected from urinary gDNA.2
Nonetheless, nucleic acid quantity and quality can be compromised from these starting materials reducing their compatibility with all NGS applications. The challenge then arises when choosing a suitable NGS workflow as, and it cannot be over-emphasized enough, the quality of your sequencing data is entirely down to the quality of the DNA that goes into library preparation.
1- Methods to assess quality and quantity characteristics of DNA
Two specific measurements, to determine DNA suitability will be specified in NGS library preparation protocols. They need to be conducted ahead of time and are the same regardless of your DNA type:
- Quantitative value, i.e., DNA concentration (ng/µl)
- Qualitative value, i.e., DNA integrity number (DIN) and % fragments within a specific size (bp) range
Note that quantity and quality assessments are not restricted to initial DNA assessment and are necessary throughout library preparation quality-check stages, as well as when calculating the final library concentration (nM).
Quantitative assessment – determining DNA quantity
NGS assays have validated sample input ranges and working outside these values could impact the sensitivity and accuracy of your data, therefore exact quantification is vital.
Fluorometry is the gold standard for obtaining DNA concentration and uses an intercalating fluorescent dye, PicoGreen, that binds to double-stranded DNA (dsDNA). The PicoGreen/DNA specificity is such that background contamination is ignored preventing an over estimation in concentration. Single-sample platforms, such as the Qubit are commonplace, as are plate readers for handling high sample throughput.
A second platform often used for obtaining DNA concentration is the NanoDrop spectrophotometer. In relation to NGS this is not a recommended method, but we are mentioning it as many laboratories have this in place. It works on the principle of measuring the concentration of compounds by their UV light absorption patterns, with dsDNA detectable by a visual peak at 260 nm.
Concentration values can be prone to inaccuracy, especially at low DNA concentrations, if other nucleic acid types are present (RNA or single-stranded DNA) or contaminants, such as phenol and ethanol, carried over from the extraction process. Contamination levels can be determined by assessing the 260:230 nm and 260:280 nm ratios and recording this is useful for troubleshooting as agents carried over can affect downstream genomic applications.
Qualitative assessment – determining DNA integrity
Ahead of library preparation, the degree of DNA fragmentation needs assessing as NGS protocols will specify their validated integrity values.
Why?
- It can affect assay input recommendations
- Determine if fragmentation is required
- Confirm the type of NGS workflow open to you. For instance, if your DNA is highly fragmented then it won’t be suitable for whole genome sequencing, so you need to explore what options are open to you instead ie, custom panels.
When determining sample integrity two platforms dominate the market; the 2100 Bioanalyzer and TapeStation. Both technologies are based on specific binding of fluorescent dye to nucleic acid and an output provides sample size (bp), quantity (ng/µl), and integrity.
The values are calculated against a set of size standards, and you can see the results in either gel format, or as an electropherogram. The degree of fragmentation is be defined as:
- The DNA Integrity Number (DIN)
- Percentage of fragments within a defined size range
2- DNA sample qualitative characteristics
Below we show the profiles you can expect from the TapeStation for gDNA, FFPE DNA and cfDNA.
Genomic DNA (gDNA)
Extracted from: liquid and tissue biopsies ie, blood, saliva, urine, CSF and fresh/frozen tissue
Considerations:
- Quantity: dependent on amount of material but high yields are usual especially from blood and saliva
- Quality: gDNA is unfragmented with a high molecular weight >60kb. DIN values >8.5
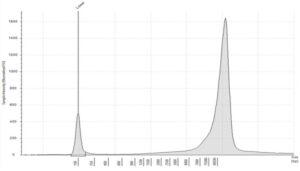
Figure 2: Typical profile of high molecular weight gDNA, from tissue and liquid biopsies, analyzed on the Agilent Genomic DNA ScreenTape, DIN 9.2 (image courtesy of Informed Genomics)
Formalin-fixed paraffin-embedded DNA (FFPE)
Extracted from: Tissue specimens that have gone through a fixation process embedding them in paraffin wax blocks. These blocks are sliced into scrolls, or placed onto glass slides, and the DNA can be extracted at any time
Considerations:
- Quantity: This is reliant on the amount of tissue present in the FFPE scroll/slide
- Quality: Highly fragmented due to the formalin fixation process.3 DIN values indicate the level of fragmentation4
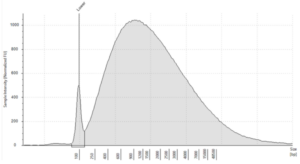
Figure 3: Typical electropherogram profile from FFPE DNA on an Agilent Genomic ScreenTape, DIN 2.5) (image courtesy of Informed Genomics)
Cell-free DNA (cfDNA)
Extracted from: Bodily fluids, such as saliva, plasma, serum, CSF and urine
Considerations:
- Quantity: cfDNA is present at low concentrations averaging 1.8-4.4 ng/ml in healthy individuals, whereas for cancer patients it can be considerably higher at ~180 ng/ml5
- Quantity: Circulating tumor DNA (ctDNA) is a component of cfDNA originating from proliferating tumor cells present in serum and plasma.6 It and can be as little as 0.01% of the cfDNA fraction with concentration correlating with tumor size and stage.6 This makes tumor derived genetic variants difficult to detect due to high background cfDNA levels
- Quality: Fragments ranging between 120-220 bp, represent repeated nucleosome positions (mono-nucleosome, di-nucleosome and tri-nucleosome) reflecting nucleosome repeats at regular intervals. There will be a maximum peak at ~167 bp. Fragmentation pattern can vary, and it is now accepted that circulating tumor DNA (ctDNA) and fetal-derived DNA (cffDNA) commonly exhibit higher fragmentation than the cfDNA shed by non-neoplastic or maternal tissues, respectively6
- Quality: the Agilent TapeStation Cell-free DNA ScreenTape does not generate a DIN value, but auto generates the % of fragments between 50-700 bp
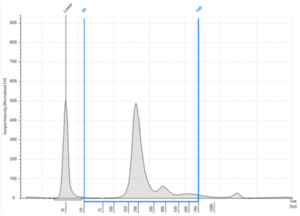
Figure 4: Typical cfDNA profile analyzed on an Agilent Cell-free DNA ScreenTape showing % cfDNA as 93.25% (50-700 bp region analysis) (image courtesy of Informed Genomics)
Note that, although the concentration values will be calculated on these platforms, the Qubit is the more reliable of the two methods for quantity assessment, so it is important to use a fluorometer for quantity, and TapeStation/Bioanalyser for sizing.
3- How DNA quantitative and qualitative characteristics influence the choice of NGS assay
So far, we have discussed DNA type and their unique characteristics. For this final section we will investigate sequencing options, and their compatibility with DNA type. Due to the variety of NGS methods commercially available, and the amounts of complex data generated, it can be overwhelming to choose.
What are the three NGS assay workflows for DNA sequencing?
Consider the following questions to help narrow the choice of NGS workflow:
What is your research/clinical question? Eg, finding novel germline variants from patients within a subset of the population? Or detecting the presence of specific cancer genes from a liquid biopsy?
- What type of NGS assay is suitable for answering the question above? Gene targeted panel, or a comprehensive approach?
- What type of starting material do you have? Liquid biopsy, tissue or FFPE slides/scrolls?
- What type of DNA sample can you expect from your extraction material? gDNA, FFPE or cfDNA?
- Will your extracted DNA type be suitable for providing the quality of data you need to answer your question?
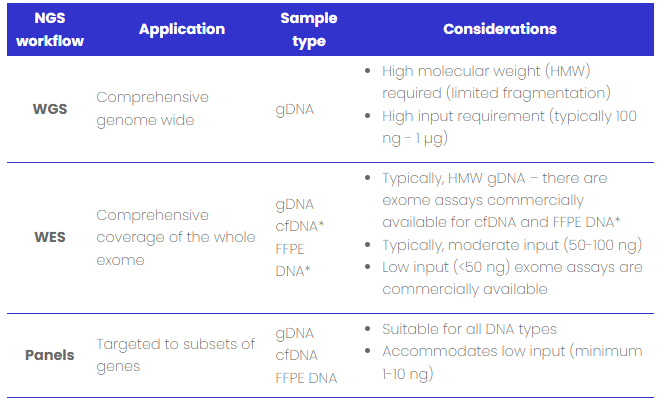
Table 1: DNA sample type compatibility with NGS workflows
Conclusion
We hope you now feel in a position to start planning your NGS project. Here at Nonacus we can help you with this task as our Cell3™ Technology NGS library preparation kits, and targeted panels, are compatible with gDNA, cfDNA and FFPE DNA. We can also help you design your own targeted panel, so to learn more about this, and how we can support you in your research and clinical applications visit our, website:
- Clinically validated workflows including bioinformatics
- Whole exome sequencing (human)
- Whole exome with additional clinical content - Cell3 Target Nexome
- Targeted panels covering pre-natal, germline and oncology disease
- Unlock the power of precision NGS; design your own targeted gene panel using our in-house Panel Design Tool
References:
1. Behjati S, Tarpey PS. What is next generation sequencing? Arch Dis Child Educ Pract Ed. 2013;98(6):236-238. doi:10.1136/archdischild-2013-304340
2. Ward DG, Gordon NS, Boucher RH, et al. Targeted deep sequencing of urothelial bladder cancers and associated urinary DNA: a 23-gene panel with utility for non-invasive diagnosis and risk stratification. BJU Int. 2019;124(3):532-544. doi:10.1111/BJU.14808
3. Ohmomo H, Komaki S, Ono K, et al. Evaluation of clinical formalin-fixed paraffin-embedded tissue quality for targeted-bisulfite sequencing. Pathol Int. 2021;71(2):135-140. doi:10.1111/pin.13054
4. Hiramatsu K, Matsuda C, Masago K, et al. Diagnostic utility of DNA integrity number as an indicator of sufficient DNA quality in next-generation sequencing–based genomic profiling. Am J Clin Pathol. 2023;160(3):261-267. doi:10.1093/ajcp/aqad046
5. Lee H, Park C, Na W, Park KH, Shin S. Precision cell-free DNA extraction for liquid biopsy by integrated microfluidics. doi:10.1038/s41698-019-0107-0
6. Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. doi:10.1186/s40246-019-0220-8
7. Whole genome sequencing — Knowledge Hub. Accessed October 6, 2023.
8. Exome. Accessed October 6, 2023.
9. Gene panel sequencing — Knowledge Hub. Accessed October 6, 2023.