Let’s Go Dotty for Lynch Syndrome – bridging the gap between testing and diagnosis
By Celina Whalley on March 22nd, 2024 | Reviewed by Victoria Simms
Finding the missing 95%
Unfortunately, 95% of people with Lynch Syndrome, also known as hereditary nonpolyposis colorectal cancer (HNPCC) syndrome, are unaware they have a pre-disposition to different cancer types.1 This is a large number of people who, if screened and found positive, would benefit from cancer risk management, early diagnosis and treatment to ultimately improve their outcome. In the UK, the gap between testing and diagnosis of Lynch Syndrome sparked the initiative to set up Let's go dotty: Lynch Syndrome Awareness Day held today on March 22nd, 2024. The drive behind this campaign is to highlight why testing for Lynch Syndrome, especially in families following cancer diagnosis, is so important whilst supporting the clear ambition of the NHS to prevent cancer wherever possible.
One test for Lynch Syndrome
Current predictive testing strategies for Lynch Syndrome focus on variant detection within in the mismatch repair genes MLH1, MSH2, MSH6, PMS2 and EPCAM. It is commonplace to take a multi-strategic approach, like immunohistochemistry (IHC) and microsatellite instability (MSI) testing, followed by further hyper-methylation and germline verification to reach a diagnostic conclusion.2,3
Consequently, turn-around times, and costs, are increased when multiple pathways are required, but there is a next generation sequencing (NGS) single-test solution for Lynch Syndrome screening. GALEAS™ Hereditary Plus detects SNVs in all the key Lynch Syndrome associated genes so there is no need for a multiple testing strategy to determine these variants, and the workflow can be implemented into any genetic testing laboratory.

Figure 1: Showing the laboratory workflow of GALEAS HereditaryPlus
Learn more about GALEAS HereditaryPlus
Overcoming challenges in SNV calling within homopolymers with GALEAS HereditaryPlus
The bioinformatic pipeline callers and panel design have been co-developed enabling single nucleotide variation (SNV) detection in homopolymers characteristic to Lynch Syndrome MMR genes. These stretches of consecutive identical bases are a common source of sequencing error. To evaluate the stringency of the assay Lynch Syndrome positive samples were included during the development of the panel, working in collaboration with the West Midlands Regional Genetics Laboratory (WMRGL).
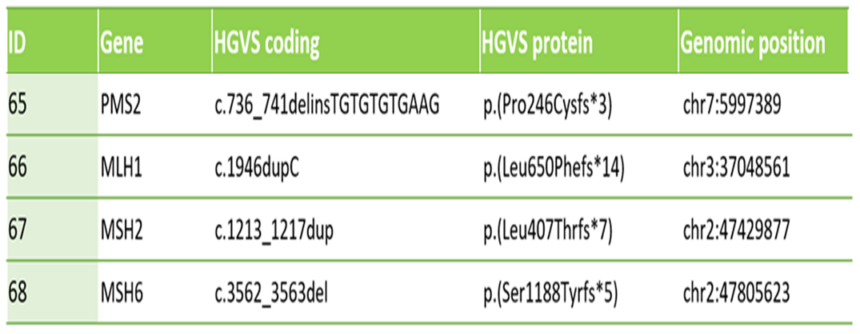
Table 1: Shows clinically relevant deletion and duplications in the major Lynch Syndrome associated genes called by GALEAS HereditaryPlus.
As part of their routine hereditary cancer screening workflow WMRGL analyzed these samples for variants in MLH1, MSH2, MSH6 and PMS2. Comparing their data, against the results from GALEAS HereditaryPlus , 100% concordance was observed confirming how robust and dependable this NGS assay is as a diagnostic tool for Lynch Syndrome.
“We were blown away by the Nonacus GALEAS HereditaryPlus data and its concordance”. Samantha Butler, Principal Clinical Scientist, NHS WMRGS, Birmingham, UK.
Reducing time between testing and cancer diagnosis
Benefits to using GALEAS HereditaryPlus
- Shorter turn around times; one assay for routine hereditary cancer screening alongside Lynch Syndrome testing
- Reduced need for additional costly MLPA assays to obtain CNV information
- Adaptable gene content to meet testing guidelines
The goal: working towards disease prevention
To support commitments towards cancer prevention targets GALEAS HereditaryPlus provides a cost effective, single genetic testing solution, with data analysis, for diagnosing Lynch Syndrome, alongside other hereditary cancers. Closing the time between testing and diagnosis of Lynch Syndrome ultimately adds to the welfare of patients and their families. Be involved in minimizing this gap by supporting Lynch Syndrome Awareness Day on March 22nd.
We fully support laboratories in implementing GALEAS HereditaryPlus into their hereditary cancer screening workflows. To learn how we can help you do the same, contact us at Nonacus.
References
1- Lynch Syndrome. https://southeastgenomics.nhs.uk/lynch-syndrome/Accessed March 12th, 2024
2- Implementing Lynch syndrome testing and surveillance pathways A handbook to support local systems https://www.england.nhs.uk/wp-content/uploads/2021/07/B0622-Implementing-Lynch-syndrome-testing-and-surveillance-pathways-version-1.2.pdf, Accessed March 12th, 2024.
3- Seppälä TT, Latchford A, Negoi I, Sampaio Soares A, Jimenez‐Rodriguez R, Sánchez‐Guillén L, et al. European guidelines from the EHTG and ESCP for Lynch syndrome: an updated third edition of the Mallorca guidelines based on gene and gender. British Journal of Surgery. 2021;108(5):484-98.
