The NEW sample-to-report NGS bladder cancer test: GALEAS Bladder – All you need to know
By Victoria Simms on September 20, 2023 | Reviewed by Celina Whalley

Bladder cancer is currently one of the most difficult cancer types to detect and treat. However, when diagnosed early, this disease is highly treatable with over 90% of patients surviving 5 or more years.1 At present, over 600,000 people are diagnosed with bladder cancer each year making it the tenth most common cancer type worldwide.2
Along with being challenging to detect due to a lack of obvious specific symptoms,3 bladder cancer is one of the most expensive cancer types to diagnose.4 The current method relies on an in-clinic flexible cystoscopy procedure to visually inspect the bladder. This is done via insertion of a camera into the urethra to look for any signs of abnormalities or malignancies. Shockingly, 88% of these procedures turn out to be unnecessary, meaning that patients are being subjected to a needless, invasive, and uncomfortable procedure, and one that is putting strain on healthcare resources and burdening cystoscopy clinics.5
To improve bladder cancer management for both patients and clinicians, Nonacus developed and launched GALEAS Bladder earlier this year. This urine-based molecular biomarker test is highly accurate and sensitive at detecting all stages and grades of bladder cancer. It provides a simple sample-to-report, molecular triage for patients with hematuria; streamlining the clinical diagnosis process and reducing the need for cystoscopies.
In this blog, we answer some of the frequently asked questions from clinical laboratories looking to offer GALEAS Bladder as a test.
Question 1: How can GALEAS Bladder enhance the current standard of care pathway for patients with bladder cancer?
The current standard of care pathway for patients with hematuria (blood in urine) is to undergo a flexible cystoscopy procedure. This requires an in clinic appointment and the procedure is invasive and uncomfortable for the patient. It also costs the healthcare provider both time and money. In the UK alone, flexible cystoscopy costs the NHS £88 million per year. In addition, the sensitivity and specificity of cystoscopy is operator dependent and around 88% of procedures could be avoided.
In contrast, GALEAS Bladder requires no hospital visit as the patient only needs to provide a urine sample, which can be collected using a simple collection device provided as part of the test, in the comfort of their own home. This reduces the burden in cystoscopy clinics and provides a far more comfortable and convenient experience for patients.
“I’ve had a few cystoscopies as part of my ongoing monitoring and they’re not something I look forward to. If instead of that I just had to give a urine sample that would be a massive, positive change. I really hope this test becomes part of standard treatment soon.” -NHS Bladder Cancer patient from Cancer Research UK.
Question 2: What is the sensitivity and specificity of GALEAS Bladder and how does this compare to the current gold standard of flexible cystoscopy?
This new test can detect all stages and grades of bladder cancer and by covering such a wide range of markers, GALEAS bladder offers the potential to be used across the whole bladder cancer care pathway; from hematuria triage, patient monitoring, detecting relapse, minimal residual disease (MRD) surveillance as well as companion diagnostics.
GALEAS Bladder has equivalent sensitivity and specificity to cystoscopy across all grades and stages of bladder cancer (see Figure 1).
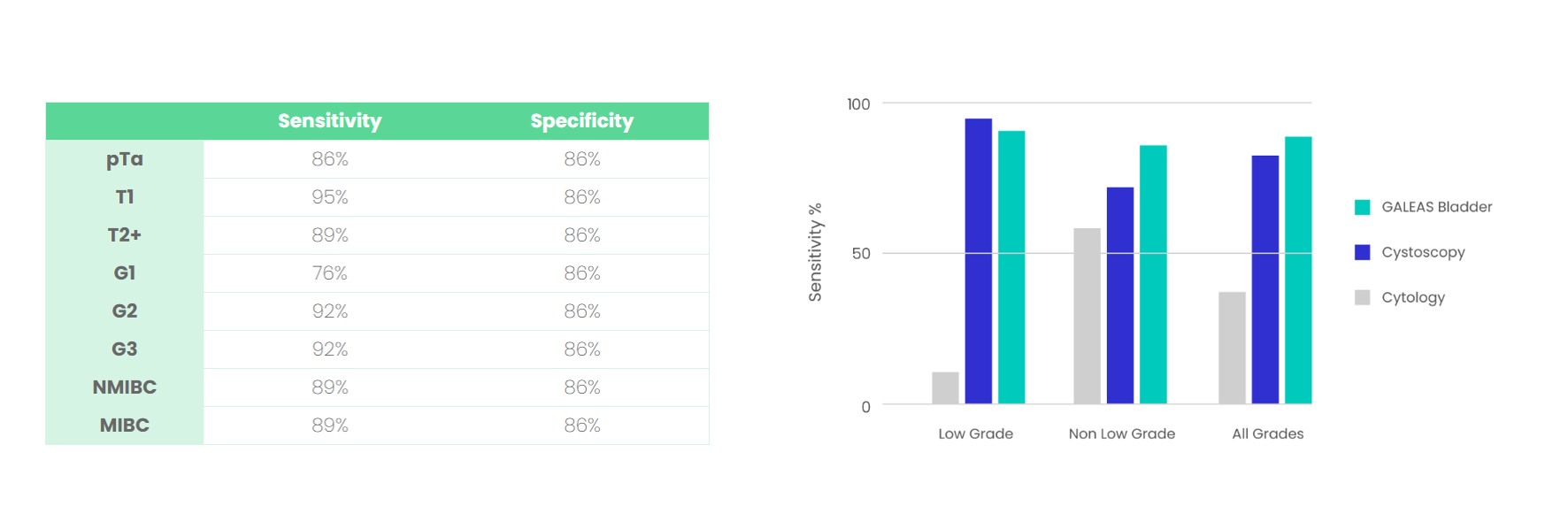
Figure 1: GALEAS Bladder performance across all stages of bladder cancer.
Question 3: What sort of molecular test is GALEAS Bladder and what does the test consist of?
GALEAS Bladder is an NGS based test. It consists of four components:
- Urine collection device which is shipped ready for dispatch to patients. It is a simple, intuitive cardboard collection device, which allows patients to provide a urine sample in the comfort of their own home. It is bar-coded for full traceability
- High-throughput genomic DNA extraction kit which extracts tumor-derived DNA from urinary cell-pellets using an easily scalable and automatable magnetic bead-based protocol
- Comprehensive gene panel including 451 somatic mutations found across 96% of bladder cancers
- Cloud-based analysis software delivers a simple positive or negative result for the likely presence of bladder cancer.
Question 4: How was the test validated?
770 patient urine samples from three UK clinical cohorts collected from hematuria clinics were used in the validation study.
GALEAS Bladder was developed in collaboration with Prof. Rik Bryan and Dr. Doug Ward from the University of Birmingham UK. They presented all the details of the test design and validation process in a recent webinar, which you can watch on-demand now: Validation of a Non-Invasive Urine-Based NGS Bladder Cancer Test webinar.
Question 5: What genes are tested for using GALEAS Bladder?
The NGS sequencing panel targets promoter and exonic regions of 23 of the most relevant genes associated with bladder cancer (see Table 1). These have been identified by a combination of publicly available data and deep exon sequencing.
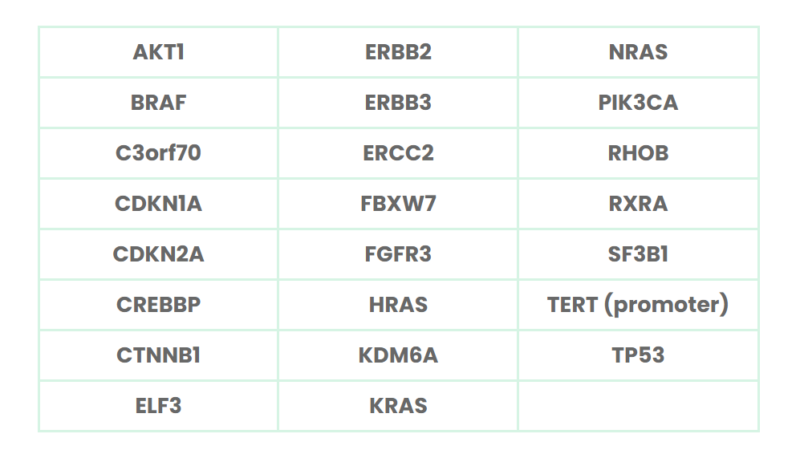
Table 1: Gene list for GALEAS Bladder. Variants in these genes covered 96% of all stages and grades of bladder cancer cases.
Question 6: What volume of urine is required from the patient to perform the test?
The patient will need to provide between 30-50 mls of urine for the test, this can be collected in the comfort of their own home.
Question 7: Are there any incidences, medications or treatments that prevents the test from being applicable for the patient?
For the use in the hematuria setting, there are no medications or treatments that we are aware of which could impact the use of GALEAS Bladder, other than the ability of the patient being able to provide a urine sample.
Question 8: How much genomic DNA is required to perform the test?
The test is optimized for 20 ng of cell pellet genomic DNA.
Question 9: Could I use cell-free DNA as well as cell pellet genomic DNA in the test?
Yes, it should be possible to use cfDNA as well as cell pellet genomic DNA. GALEAS Bladder has been tested and validated using cpDNA, as in the majority of cases the cpDNA will be of higher quantity compared with the cfDNA that is obtained from a urine sample. Therefore it is likely to be easier and more accurate to perform the test using cpDNA.
Question 10: Can I use a sequencing platform other than Illumina for the GALEAS Bladder test?
The test has been validated on Illumina sequencing chemistry, however the test could feasibly be converted to other sequencers. Please get in touch if you wish to use a sequencer other than Illumina.
Question 11: Is the GALEAS Bladder analysis software available as a desktop version?
No, our analysis software is cloud based with Amazon Web Services.
Question 12: Where is the GALEAS Bladder analysis software hosted, given that it is cloud based?
Currently, the GALEAS software is hosted in UK cloud data-centres, with plans for expansion across different geographies.
Question 13: What is the planned retention period on GALEAS Bladder for analysis files?
The general retention policy is as follows.
Patient sample data will be retained and available on-line on the following basis;
- Individual FASTQ for a patient and any resultant report, BED, BAM and VCF’s for a period of 1 year from the point of successful processing.
- Longitudinal Monitoring ( where a patient will have multiple samples over a period of time) for FASTQ’s and resultant report, BED, BAM and VCF’s for a period of three years from the date of the last successful sample processing.
After this time, all data (both input FASTQ’s and any resultant outputs from the GALEAS software) will be held in ‘cold storage’ for a period of five years. Retrieval times for this data, should the need arise, will be 10 working days. After the five year period all the data (input and resultant outputs) will be deleted.
Question 14: I would like to work with you on setting this up in my clinical laboratory, how do I go about doing this?
We would be delighted to work with you in doing this. Please get in touch and one of the Nonacus team will reach out to you as soon as possible.
In addition, we are always looking for partners to provide samples to further validate our panel for use in NMIBC and MIBC surveillance as well as companion diagnostics and stratification. If you are interested in collaborating with us please do contact us.
If you are interested in adding GALEAS Bladder as a test in your clinical laboratory or would like to access it for your patients, please get in touch and one of our team will contact you.
References
-
- Bladder Cancer: Statistics | Cancer.Net Accessed 20 Sept, 2023
- World Health Organisation | .iarc.who.int/cancer-type/bladder-cancer Accessed 20 Sept, 2023
- Pacchioni G. Non-invasive detection of bladder cancer. Nature Reviews Materials. 2022:7(9):679-.
- Matuszczak M, Kiljańczyk A, Salagierski M. A liquid biopsy in bladder Cancer—the current landscape in urinary biomarkers. International Journal of Molecular Sciences. 2022;23(15):8597.
- Kelly JD, Fawcett DP, Goldberg LC. Assessment and management of non-visible haematuria in primary care. BMJ. 2009;338.