Tracking the evolution of SARS CoV-2 using whole genome sequencing (WGS)
November 3, 2021. Reviewed by Victoria Simms, April 23, 2024.
Why monitoring the evolution of the virus is crucial to public health
Viral genome sequencing: the background story
Rapid whole viral genome sequencing, decoding a pathogen's entire genome, is essential to public health organizations and epidemiologists for tracking and surveillance of disease outbreaks. Genomic epidemiologists around the world are monitoring genetic variation in pathogens to elucidate how diseases spread.
In December 2019, patients presenting with viral pneumonia were found to be epidemiologically associated with the Huanan seafood market in Wuhan, China.¹ It was the use of next generation sequencing (NGS) technology that identified the cause as SARS-CoV-2. The outbreak rapidly spread to many regions of the globe and within three months the World Health Organization had declared a global pandemic.
At Nonacus, we responded to this crisis by developing a novel and highly rapid whole genome sequencing (WGS) library preparation approach for viral genomes coupled with Oxford Nanopore Technology's sequencing platform. This development was funded by a grant from Innovate UK, the UK's innovation agency. Further, we used this funding to develop an additional, novel test that detects and sequences the whole genomes of multiple human respiratory viruses in a single test.
How did Nonacus democratize access to testing?
Our innovative method, transitioned into a service, addressing the need for widespread access to affordable and rapid SARS-CoV-2 WGS. Through the provision of cost effective and sensitive extraction kits, and CE-IVD marked qRT-PCR antigen kits, to many laboratories, we were able to further support through a complimentary SARS-CoV-2 whole genome (WGS) service. By doing so, many more laboratories were able to offer testing, creating additional competition in the market place and helping to avoid monopoly.
How did Nonacus support public health initiatives using SARS-CoV-2 whole genome sequencing (WGS)?
WGS of the SARS-CoV-2 virus revealed information on the pathogen's structure and function. To survive and spread, the virus replicates within and between individuals. During this inaccurate process the virus accumulates small variations in its genome. Genomic epidemiologists track these genetic changes to monitor how and where the disease is spreading. By sequencing the genome of SARS-CoV-2, in patients with COVID-19 in its entirety, genetic changes can be detected that arise in the virus as it spreads through the population. WGS increased our understanding of the changes in the viral genome allowing public health institutions to monitor the development and spread of the epidemic, guide control measures, and inform vaccine research.² The three main aims in tracking viral genetic variation covered:
1) To understand how the virus is transmitted
Understanding variation in the genetic sequence of the SARS-CoV-2 viral genomes collected from patients allows monitoring of how Covid-19 spreads between individuals over time. It may also help to identify hotspots to inform and guide control procedures.
2) To design vital lifesaving treatments and vaccines
WGS informs the design of anti-viral drug treatments and vaccines that target specific features of the virus. It also reveals insight in how drug and vaccine effectiveness might change through viral evolution.
3) Monitor viral evolution
Keeping track of genetic variation in the virus may serve as early warning of a more virulent virus or drug/vaccine resistant strains; supporting measures to minimize disease spread and for designing new treatments and vaccines.
What the future looked like: the pan-respiratory panel test
The annual winter influenza season undoubtedly places increased pressure on the NHS. Co-infection of Influenza and SARS-CoV-2 is known to double the mortality risk.³ The symptoms of SARS-CoV-2 infection are often difficult to distinguish from multiple respiratory pathogens. In response to this we expanded our technology to address a complex public healthcare need whereby patients presented with respiratory illness without a clear diagnosis. Our pan-respiratory viral enrichment panel approach leveraged Oxford Nanopore's WGS platform and had the power to detect multiple respiratory pathogens in one rapid test. This test identified patients with co-infections so was particularly relevant through the winter season.
The pan-respiratory panel test allowed detection and complete WGS of the following respiratory pathogens in a single process:
- Coronavirus - All Human
- Influenza - All A and B
- Metapneumovirus
- Rhinovirus - A, B, C and Enteroviruses
- Parainfluenza Virus - 1,2,3,4
- RSV - A and B
- Measles
- Mumps
- Rubella
The pan-respiratory panel method
The pan-respiratory panel test had application for both Oxford Nanopore Technology long read sequencing and Illumina short read sequencing approaches. The flow diagram (Figure 1) shows the streamlined workflow from nucleic acid extraction to sequencing data generation.
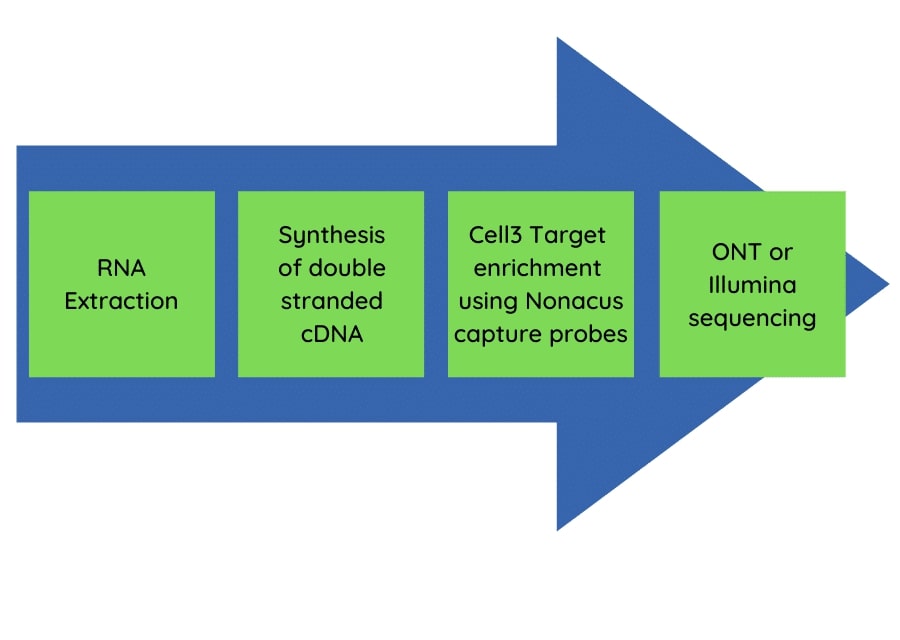
Figure 1: The pan-respiratory panel method workflow.
References
1. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine. 2020; 382(8):727-33
2. Gardy JL, Loman NJ. Towards a genomics-informed, real-time, global pathogen surveillance system. Nature Reviews Genetics. 2018;19(1):9-20.
3. Lacobucci G. Covid-19: Risk of death more than doubled in people who also had flu, 2020. BMJ 370 m:3720. DOI: 10.1136/bmj.m3720.