Validated for FFPE and ctDNA samples
Developed for and validated on ctDNA, gDNA and FFPE DNA for oncology applications like early cancer detection and monitoring cancer post treatment.
Confidently call low frequency variants
QC validation guarantees uniformity of coverage which combined with error suppression technology ensures confident calling of all mutations down to 0.1% VAF.
Simple and rapid custom design process
Our online panel design tool means you can design and optimize a panel in minutes and our rapid manufacturing process means minimal time between design and delivery.
Optimized for small and large panels
Cell3 Target enrichment has been optimized to deliver efficient target capture and is unrivalled on target performance regardless of panel size from 1-1000 genes.
Targeted resequencing panels for low frequency mutation detection
While detection of low frequency mutations <20% VAF has been possible with NGS methods for a number of years, accurate detection of ultra-low frequency mutations (below 5% VAF) has been hampered by excessive false positives, or artifactual, variant calls.
The need to accurately call variants below 5% VAF is important for the advancement of non-invasive techniques like liquid biopsy testing where the sample used, circulating tumor DNA (ctDNA), is in low abundance and requires ultra-sensitive sequencing methods to detect it.
Targeted sequencing through the use of custom designed NGS panels offers a cost-effective method for rare variant detection. Without the data burden of whole genome sequencing (WGS) or whole exome sequencing (WES) it allows researchers to achieve very high depths of coverage providing the increased sensitivity and accuracy needed for ctDNA analysis. However, deep sequencing is not without its flaws as sequencing low-abundance DNA at high depth also increases the level of sequencing artifacts, creating background noise and potentially masking low-frequency variants. In light of this, it is essential when you are working with ctDNA that extra care is taken to ensure that errors are suppressed, and only high-quality variant calls are retained.
Cell3 Target is a novel target enrichment system developed by Nonacus for converting any type of DNA (cfDNA, gDNA, FFPE DNA) into libraries for next generation sequencing. It uses error suppression technology to ensure confident calling of all mutations down to 0.1% VAF and is ideal for rare variant detection in liquid or tissue biopsies.
Reliable target enrichment for solid and liquid biopsy
We understand that our customers have different questions to answer so Cell3 Target has been developed for and validated on ctDNA, gDNA and FFPE DNA for a range of oncology applications, so that whether you are investigating early cancer detection, diagnosis and treatment selection, tumor heterogeneity or using ctDNA for tumor monitoring purposes, you know you will be able to use the same method and panel design.

Quick and easy workflow for FFPE, FF and ctDNA
Cell3 Target offers quick, Covaris free, enzymatic shearing for FFPE/ fresh frozen (FF) DNA samples, and is available without enzymatic fragmentation reagents for ctDNA.
- Quick and easy workflow takes less than 10 hours, with less than 2 hours hands-on time
- Supports manual or automated preparation of 1–96 samples in a single batch
- 384 patient/sample indexes ensure that the customer can use Cell3 Target on the smallest to the largest output sequencers
- Suitable for use with as little as 1 ng of genomic or ctDNA and 10 ng of FFPE
Error suppression technology for ultra-sensitive variant detection
Cell3 Target enrichments incorporate error suppression technology including unique molecular indexes (UMIs) and unique dual indexes (UDIs) which remove both PCR and sequencing errors and index hopping events. This error suppression technique, combined with our excellent uniformity of coverage, allows you to confidently and accurately call variants down to 0.1% VAF and enables generation of sequencing libraries from as little as 1 ng of cfDNA input. We provide advice and provision of ready to go analysis scripts for error removal using UMIs.
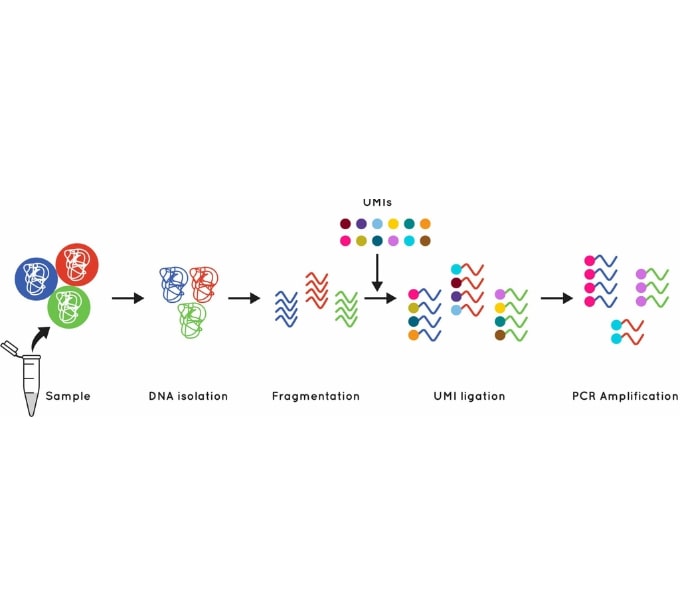
Figure 1: Using UMIs to identify and quantify individual DNA molecules during library preparation increases sensitivity.

Successful panel design for liquid or solid biopsy
Good design is key to a successful NGS panel ensuring adequate coverage and robust variant calling. It’s not always easy to achieve, especially the first time, but our Panel Design tool makes it simple and easy allowing you to optimize panels in minutes. Create designs instantly from scratch using uploaded BED files, gene lists or genomic coordinates, customize catalog products or add your own content onto one of our fixed panels.
- Easily merge, compare and share panels
- Get coverage data instantly and request a quote
- Rapid manufacturing process ensures minimal time between design and delivery
Uniformity of enrichment optimizes sequencing performance
The baits used in Cell3 Target panels are designed to deliver excellent uniformity of coverage. By improving uniformity of coverage and reducing the number of low coverage exons, our targeted enrichment optimizes sequencing efficiency and sample capacity per sequencing run ultimately reducing your sequencing costs.
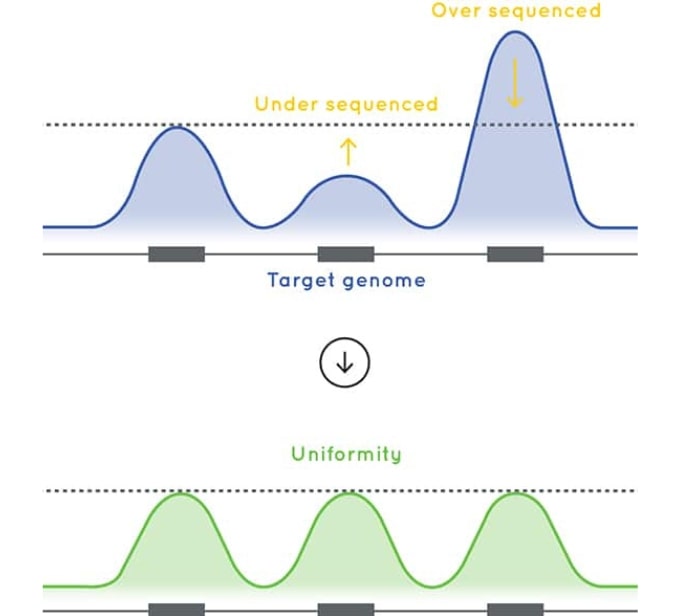
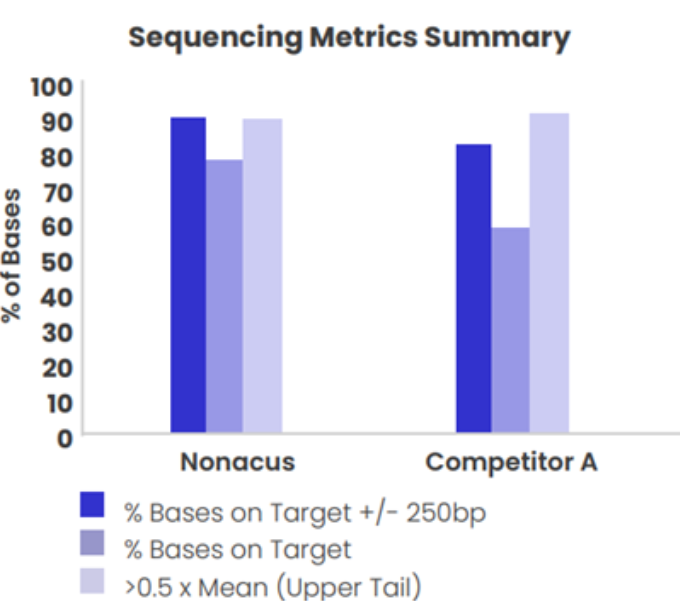
Figure 2: A comparison of coverage uniformity and % target capture for a 40 Kb panel designed using Cell3 Target technology and Company (A) High Sensitivity enrichment kit. Targeted enrichment performed on genomic DNA samples and sequenced on the Illumina MiSeq.
Improved on-target for small and large panels
Most capture target enrichment technologies suffer from a significant percentage of off target sequencing reads when targeting small regions of the genome. Cell3 Target has been developed so that whether you wish to sequence 1 or 1000 genes you will find lower off target and more uniform coverage when compared with alternative capture technologies.
See our customer publications
Cell3™ Target panels are available with one of two versions of our library preparation kits:
- Fragmentation: for use with gDNA (FF or FFPE)
- Non-Fragmentation: for use with cell-free DNA
| Panel Size - XS | 50-500 probes | |
| Frag | Non-Frag | |
| 48 Samples | NGS_C3C_XS_FR_48 | NGS_C3C_XS_NF_48 |
| 96 Samples | NGS_C3C_XS_FR_96_A/B/C/D* | NGS_C3C_XS_NF_96_A/B/C/D* |
| Panel Size - S | 501-1000 probes | |
| Frag | Non-Frag | |
| 48 Samples | NGS_C3C_S_FR_48 | NGS_C3C_S_NF_48 |
| 96 Samples | NGS_C3C_S_FR_96_A/B/C/D* | NGS_C3C_S_NF_96_A/B/C/D* |
| Panel Size - M | 1001-5000 probes | |
| Frag | Non-Frag | |
| 48 Samples | NGS_C3C_M_FR_48 | NGS_C3C_M_NF_48 |
| 96 Samples | NGS_C3C_M_FR_96_A/B/C/D* | NGS_C3C_M_NF_96_A/B/C/D* |
| Panel Size - L | 5001-7500 probes | |
| Frag | Non-Frag | |
| 48 Samples | NGS_C3C_L_FR_48 | NGS_C3C_L_NF_48 |
| 96 Samples | NGS_C3C_L_FR_96_A/B/C/D* | NGS_C3C_L_NF_96_A/B/C/D* |
| Panel Size - XL | 7501-10,000 probes | |
| Frag | Non-Frag | |
| 48 Samples | NGS_C3C_XL_FR_48 | NGS_C3C_XL_NF_48 |
| 96 Samples | NGS_C3C_XL_FR_96_A/B/C/D* | NGS_C3C_XL_NF_96_A/B/C/D* |
| Panel Size - XXL | 10,000-25,000 probes | |
| Frag | Non-Frag | |
| 48 Samples | NGS_C3C_XXL_FR_48 | NGS_C3C_XXL_NF_48 |
| 96 Samples | NGS_C3C_XXL_FR_96_A/B/C/D* | NGS_C3C_XXL_NF_96_A/B/C/D* |
* To provide flexibility in multiplexing samples, our 96-sample kits offer a choice in adapter plate:
A = Adapter plate with indexes 1-96
B = Adapter plate with indexes 97-192
C = Adapter plate with indexes 193-288
D = Adapter plate with indexes 289-384
