Cell3 Preserver Tube
They have a three month shelf life once the pouch has been opened. Store at room temperature, 18-25oC.
The Cell3 Preserver Tubes are designed to stabilize whole blood for up to 15 days at a range of storage temperatures (4°C–37°C).
Yes, you may see some pink coloration to the plasma. The Cell3 Preserver Tubes contain stabilization additives which protect and prevent white blood cells from breaking apart. Keeping the white blood cells intact prevents genomic DNA contamination in the plasma sample.
However, red blood cells have thinner walls and may break down in a process called hemolysis. Red blood cells do not contain DNA and therefore will not contribute to any genomic DNA contamination in the plasma sample.
Bead Xtract cfDNA Kit
Plasma can be stored at -20°C for long term storage, and we recommend using DNA LoBind tubes for this purpose. Prior to cell-free DNA extraction, the plasma sample should be thawed at room temperature or in a heat block or water bath at 37°C.
Eluted DNA can be used immediately for downstream applications or stored at ≤ -20°C. We recommend using DNA LoBind tubes for storing DNA samples.
We recommend using a two-step centrifugation process to isolate the plasma, an initial spin speed of 2000 g for 10 minutes to isolate the plasma fraction from the red and white blood cells, then a second high spin of minimum 10,000 g for 10 minutes to pellet and remove intact cells and cellular debris from the plasma fraction.
During shipment or storage in cool ambient conditions, precipitates may form in some buffers. Dissolve such deposits by warming the solution at 37°C and gently shaking.
Our elution buffer consists of 10 mM Tris-HCl, pH 8.5, 0.1 mM EDTA; therefore, this low level of EDTA should not affect most downstream applications, including NGS library prep. It is perfectly fine to use water in place of the supplied buffer to elute your cell-free DNA.
If doing so, please ensure that the pH is >6.0 and that it is nuclease free. However, if you are storing your samples long term prior to library preparation, the buffer should be used and your sample quality checked before you proceed with your downstream application.
The magnetic beads need to be shaken or vortexed to fully resuspend the particles before use. Once the magnetic beads have been added to the sample, the sample must be mixed throughout the 10 minute incubation period by shaking, rocking or inverting to keep the magnetic beads resuspended in solution.
All of the Bead Xtract cfDNA Kit components are guaranteed for at least 12 months from the date of purchase when stored as follows. Magnetic Particles should be stored at 2-8°C for long-term use. Proteinase K Solution can be stored at room temperature for up to 12 months.
For longer term storage, store Proteinase K Solution at 2-8 °C. Store all other components at room temperature. During shipment or storage in cool ambient conditions, precipitates may form in some buffers. Dissolve such deposits by warming the solution at 37°C and gently shaking.
Cell3 Xtract Kit
Plasma can be stored at -20°C for long term storage, and we recommend using DNA LoBind tubes for this purpose. Prior to cell-free DNA extraction, the plasma sample should be thawed at room temperature or in a heat block or water bath at 37°C.
Eluted DNA can be used immediately for downstream applications or stored at ≤ -20°C. We recommend using DNA LoBind tubes for storing DNA samples.
We recommend using a two-step centrifugation process to isolate the plasma, an initial spin speed of 2000 g for 10 minutes to isolate the plasma fraction from the red and white blood cells, then a second high spin of minimum 10,000 g for 10 minutes to pellet and remove intact cells and cellular debris from the plasma fraction.
During shipment or storage in cool ambient conditions, precipitates may form in some buffers. Dissolve such deposits by warming the solution at 37°C and gently shaking.
Our elution buffer consists of 10 mM Tris-HCl, pH 8.5, 0.1 mM EDTA; therefore, this low level of EDTA should not affect most downstream applications, including NGS library prep. It is perfectly fine to use water in place of the supplied buffer to elute your cell-free DNA.
If doing so, please ensure that the pH is >6.0 and that it is nuclease free. However, if you are storing your samples long term prior to library preparation, the buffer should be used and your sample quality checked before you proceed with your downstream application.
No specialist equipment such as magnets or vacuum manifolds are required. You will need the following which is standard in most blood handling laboratories: water bath or heat block (55°C), microcentrifuge (capable of accommodating 1.5-2 ml tubes) and a swing bucket centrifuge (capable of accommodating 15-50 ml tubes).
Cell3 Target: Library preparation
No, you set up the sequencer the same for all applications. The 9 bp UMI is attached to the Illumina i7 8 bp index and therefore, needs 17 cycles. The i5 index has 8 bp and needs 8 cycles. Then you can choose to use them or not during your bioinformatics analysis according to the application.
UMIs are appropriate for applications that produce high levels of duplicates. For, example, when sequencing low input cfDNA to detect very low frequency variants, ultra-deep sequencing is necessary (20,000 – 30,000 raw read depth).
This results in high duplicates (~90%). When duplicates are removed using the UMI approach this will result in a consensus read depth of 2000 to 3000×. Cell3 Target’s built in UMIs enables a single workflow for all sample types and tests allowing confident and sensitive calling of mutations down to 0.1% VAF and from as little as 10 ng ctDNA input.
This allows you to choose to use them or not without any penalties in your resulting data. UMIs are not necessary when sequencing at lower depths, eg, sequencing gDNA from whole blood or from tissue samples (FFPE or FF). This is because the proportion of duplicates is much lower (for example, is approx. 5-6% when sequencing gDNA at 100× depth for panels eg, Cell3 Target Nexome panel. Please follow this link for an overview about UMIs.
Ultra-deep sequencing is a sensitive method to allow the identification of very low frequency variants. However, one caveat to this technique is the high level of duplicates generated when sequencing th same DNA fragment multiple times. A UMI (Unique Molecular Identifier) is a molecular tag consisting of a short DNA sequence that is used to identify and quantify unique DNA molecules.
These 9 bp tags, ligated to the end of DNA fragments during library preparation, enable duplicate removal. DNA molecules with identical UMIs are assumed to originate from the same initial input molecule. Sequencing reads with the same UMI are grouped together to form consensus reads. UMIs therefore, allow for PCR sequencing error correction and ultra-low frequency mutation calling. Please follow this link for an overview about UMIs.
Coverage metrics are analyzed using the covered.bed file as this file defines the genomic regions covered by one or more probes, which will extend beyond the target region. Whilst variant calling can be performed with either bed file, the target_merged.bed is considered the better choice. This is because the coverage at the edge of the covered file may be much lower.
If you intend to detect splice variants it is advisable to extend your target region at the design stage rather than rely on the extended region of the covered.bed. It is recommended that you visualize your design by loading your bed files as custom tracks in UCSC Genome Browser prior to ordering your panel design.
Only high-purity DNA samples which are free of residual salts, proteins, detergents or other contaminants should be used as input material. Library preparation can be conducted using 1–1000 ng of DNA. Fluorometric methods (such as the Qubit assay, Invitrogen) are recommended to accurately determine DNA concentration, especially when using <100 ng of DNA as input.
DNA samples should be resuspended in molecular biology grade water, a low EDTA concentration Tris-HCl buffer (such as 0.1 mM EDTA TE buffer) or a 10 mM Tris-HCl pH 8.0 saline buffer (such as QIAGEN Buffer EB or equivalent).
If DNA samples are kept in a high EDTA concentration buffer (such as 1× TE), DNA needs to be purified using a commercially available kit or DNA Purification Beads (such as Target Pure NGS clean-up beads or equivalent) and resuspended in one of the above-mentioned buffers.
A detailed workflow can be found in the Cell3 Target protocol. Please follow this link.
The fragmentation kit contains enzymatic reagents that will shear genomic DNA into fragments of comparable size to cell-free DNA. The non-fragmentation kit does not contain any enzymatic reagents, since this kit is suitable for cell free DNA, and these DNA fragments are already short in size ( ~160 bp).
The correct type of Dyna Beads is critical to the success of the capture. Only M-270 Streptavidin beads are suitable and will give the correct post capture yield. Any other beads are likely to be a different diameter and concentration and have shown to give a significant decrease in final yield.
Often, we find that the beads dry quicker than five minutes, so it is advisable to check at three minutes. Ensure that they are not shiny and try not to wait until they are cracking and overdried as beads are more difficult to resuspend and you may recover less of your sample
For best results, we recommend that you extract cfDNA and quantify just before you start the protocol. However, sometimes there is a need to extract and store cfDNA at an earlier time. In this case we advise you to store extracted cfDNA in a LoBind tube at -20˚C and quantify your samples after thawing when you are ready to start the library preparation.
We recommend the hybridization to run between 4 and 16 hours. However, you can run the hybridization overnight.
We have successfully run the protocol using cyclers that have a set maximum volume of 50 µl or 100 µl.
We do not recommend mixing Cell3 Target enrichment probes with libraries prepared from other kits because, we cannot guarantee the results. The reason being is that there would be compatibility issues since the blockers in our kits are designed for our Cell3 Target workflow.
Yes, you can purchase a combination of fragmentation and non-fragmentation kits, this is particularly useful if you would like to prepare libraries from genomic DNA alongside cell-free DNA. You can purchase combinations of kits based on sample sizes 16, 48 and 96.
A low molecular weight peak of 150-160 bp in size indicates the presence of adapter-dimers carried over from the adapter ligation reaction - as in the figure below. These should not be carried over to the final library pool. If your libraries show a significant adapter-dimer peak this may be because you have input less than 50ng of DNA into the initial reaction without diluting the adapters to 1.5 µM (1:10 dilution).
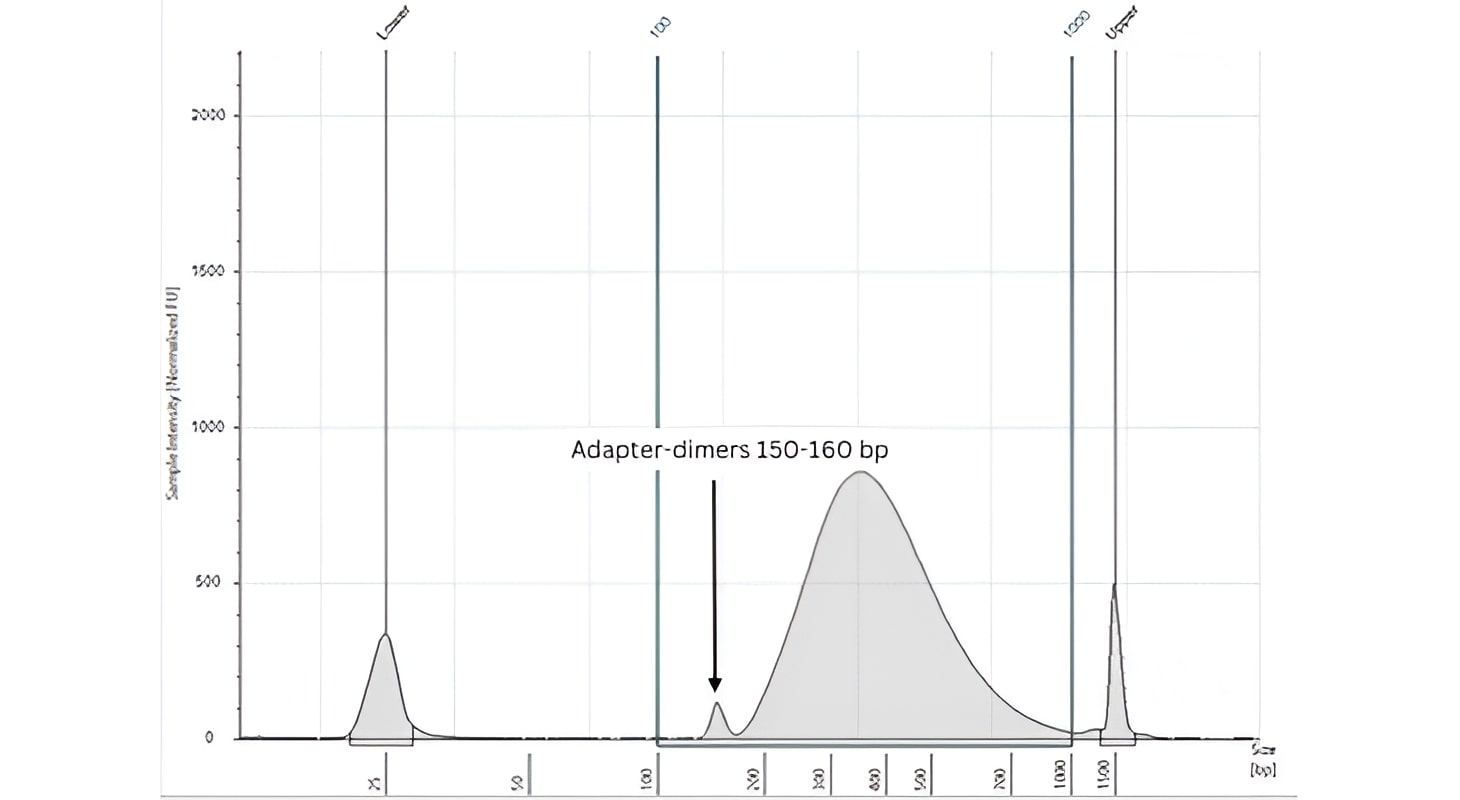
Always ensure you have accurately determined the concentration in your samples immediately prior to library preparation. You can attempt to reduce the adapter-dimer peak by performing a 0.9X (bead to sample ratio) clean-up step. However, this will result in sample loss, and you will need to perform additional PCR cycles to ensure you have sufficient material to continue. Please follow this link to see the Cell3 Target protocol document for more information.
The TapeStation profile and yield of extracted cfDNA will vary depending on the input sample. The following descriptions give examples of these:
a) cfDNA extracted from cancer patients will be a mixture of cfDNA from the normal process of apoptosis and circulating tumor DNA (ctDNA) shed from the patient’s tumor.
The amount of ctDNA shed into the blood varies with tumor type, stage and whether the patient has received treatment. There is a size difference observed in fragment length between cfDNA derived from the tumor and the patient’s normal background cfDNA.
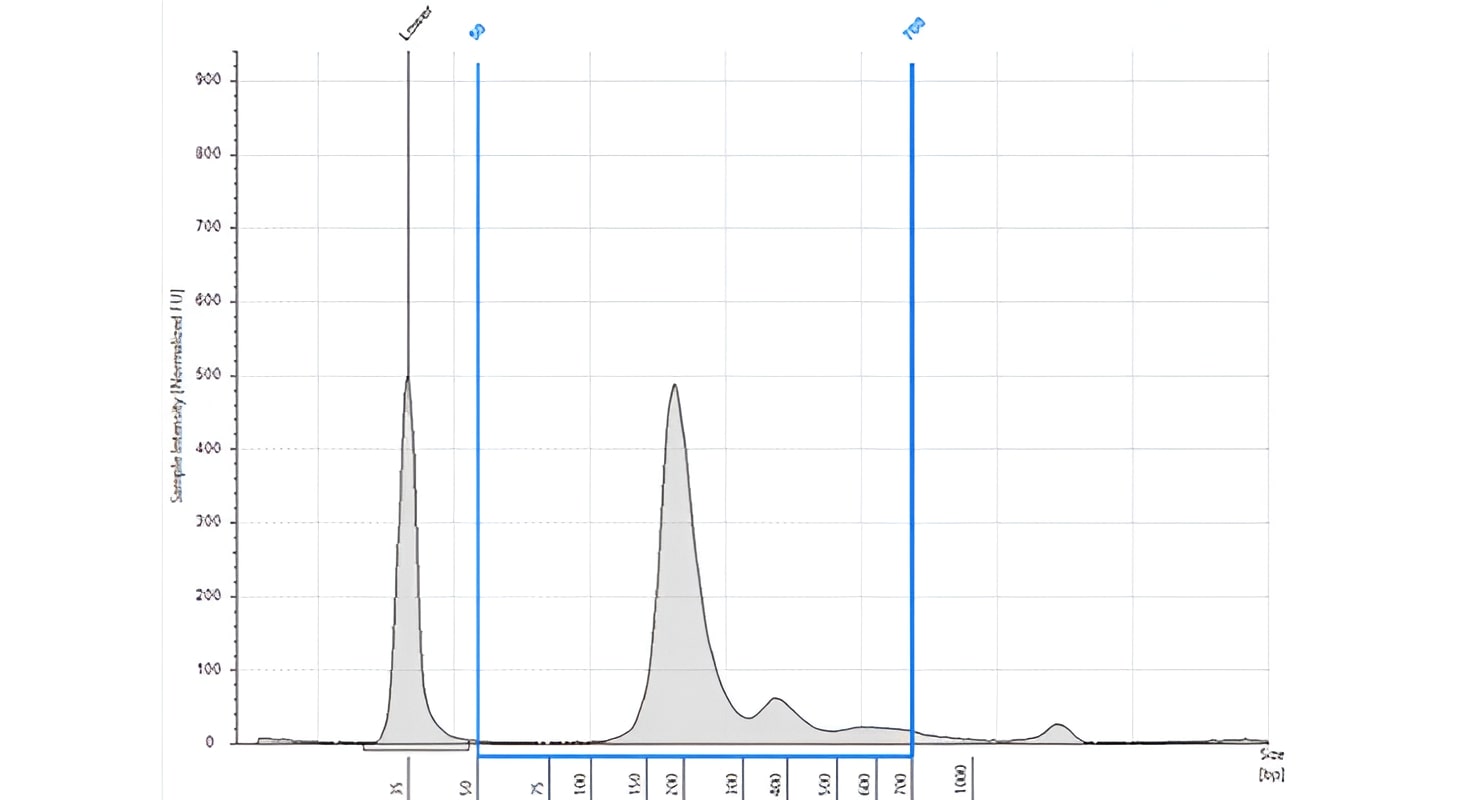
The normal cfDNA is generally around 166 bp (160-180 bp peak), which corresponds to the length of DNA wrapped around the nucleosome and is likely to be the result of normal apoptosis. Peaks in multiples of 160-180 bp are often observed. The ctDNA portion in the sample is generally a smaller peak and reveal a shorter fragment length of approx. 145 bp.
b) cfDNA extracted from patients with inflammatory conditions, infections, late-stage cancer, or transplant rejection, may reveal larger peaks of cfDNA on the TapeStation analysis.
You may see several peaks of differing sizes as multiples of 166 bp (eg, 332 bp, 498 bp, 664 bp). The figure below shows the cfDNA profile of a patient with an inflammatory condition.
There are several probable causes for this. Accurate quantification of material and dilution upfront is a primary concern if you have an under fragmented library. Additionally, please ensure that the correct time and temperature have been used for fragmentation.
If you are not using 10-100 ng and this is your first use of Cell3 Target with this sample type, we recommend first trying three different times to find the best option for your sample type and input quantity. Alternatively, there may be EDTA in your DNA extraction elution buffer.
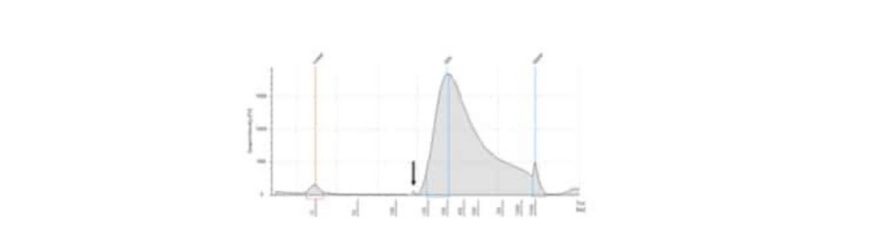
EDTA affects the enzymatic shearing and causes poor/under fragmentation. DNA stored in EDTA buffers should be cleaned up with bead or column clean up prior to use. Ideally non EDTA elution buffers such as EB buffer (10 mM Tris HCL pH 7.5-8) should be used.
Please note that the enzymatic shearing reaction should be prepared on ice and mixed adequately before incubation by pipetting or vortexing. An example of an oversized library, due to shearing issues, can be seen in the figure below and via this link to the Cell3 Target protocol.
We recommend using a fluorometric method such as the Qubit assay, Invitrogen. Additionally, you may want to perform different QC steps on your input material according to your material type.
a) FFPE derived DNA may be compromised due to formalin fixation, so we recommend performing a quality check by running your samples on a genomic screen tape (TapeStation) to determine the DIN (DNA integrity) score.
This allows you to optimize the DNA input adding more material with lower DIN scores.
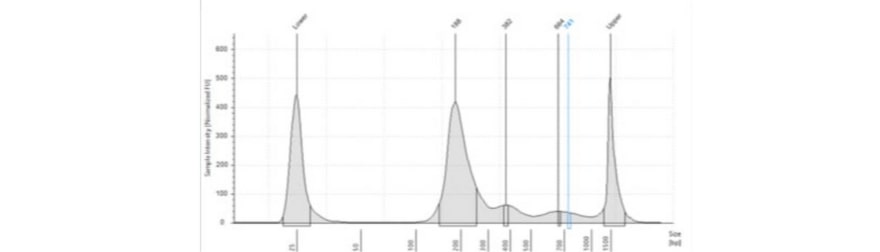
b) DNA derived from plasma may have gDNA contamination, and the extent of this can be determined using a High Sensitivity D1000 Screen Tape on the TapeStation. Contamination with gDNA can arise during the processing of plasma due to hemolysis of white blood cells.
A peak at >1500 bp is evidence of sample contamination with gDNA. You may also want to run this check to determine that the TapeStation profile reveals fragments of the expected size range. The figure below shows a Tapestation profile of cfDNA from a patient with an imflammatory condition.
Depending on the need for UMIs, the following DNA input quantities are recommended as in the Cell3 Target protocol:
(1) 1-100 ng of input DNA is recommended when wanting to take advantage of UMIs.
(2) 100-1000 ng of input DNA is recommended if UMIs are not required or when preparing PCR-free libraries. UMIs enable PCR/sequencing error removal and high accuracy single molecule counting analysis.
This is achieved by grouping identical sequencing reads containing the same UMI in a single family and creating a consensus read. Therefore, a certain amount of PCR duplicates is required to take full advantage of UMIs. However, downstream bio-informatics analysis can also be conducted ignoring the presence of UMIs.
Yes, this is possible; however, it is important to note that the library size would differ depending on the DNA input type. cfDNA is on average 166 bp in length therefore, we usually recommend 2 x 75 bp read length. For gDNA and FFPE DNA fragments are often longer, and we would recommend a 2 x 150 bp read length.
Therefore, it is important to tailor your sequencing cycle parameters to your library size so you do not sequence into your adaptors inadvertently (which would then need removing during analysis). Conversely if you sequenced your libraries at a shorter read length, this would impact the potential depth of coverage.
DNA extracted from formalin-fixed paraffin-embedded (FFPE) tissue is generally more degraded than genomic DNA extracted from fresh tissue or cells and can be chemically modified to different degrees. Depending on the level of DNA degradation, increasing quantities of input DNA need to be used during library preparation in order to achieve similar yields compared to high-purity DNA.
The DNA integrity score (or DIN score) can be determined by running FFPE DNA samples on an Agilent Genomic DNA ScreenTape (Agilent Technologies). DNA input quantities are impacted as follows: A) DIN score <3 requires 5-10 fold increase, b) DIN score 3-8, requires 1.5-4 fold increase, c) DIN score >8, requires no increase. See the Cell3 Target protocol for more information.