Tumor relevant content
Whole exome capture designed specifically for cancer research covering key intronic regions like promoters, translocations and fusions and CNVs.
Optimized performance
Delivers excellent uniformity of coverage, Cell3™ Target: Tumor Exome reduces sequencing costs and improves sample throughput.
Supports a range of sample input types
Accommodates a broad range of sample types; cell-free (cfDNA), cell-free fetal (cffDNA), and DNA from fresh frozen as well as formalin fixed paraffin embedded (FFPE) tissue.
Customizable content
Designed to be flexible, Cell3™ Target: Tumor Exome allows you to add extra content to cover targets specific to your project.
Primary tumor sequencing
We believe a different type of exome is required for cancer research. By focusing on the coding regions of the genome, whole exome sequencing offers a cost-effective solution for primary tumor profiling when compared to whole genome sequencing. It allows for deeper sequencing enabling more sensitive mutation detection and improved tumor mutation burden (TMB) calling versus smaller panels.
However, many mutation types that are known to be important in cancer, for example tumor promoters like TERT, translocations and fusions or CNVs, are not covered by standard exome enrichments.
Tumor relevant content for cancer research
The Nonacus Cell3™ Target: Tumor Exome is based on our standard Whole Exome product enhanced with baits from our Pan-Cancer panel. It includes enhanced coverage of the most clinically relevant genes from NCCN/FDA cancer treatment guidelines, 116 cancer driver genes and 345 genes in vital cancer signaling pathways.
The design, whilst exon focused, covers key intronic and promoter regions like the TERT promoter and translocations and fusions. It also contains genome-wide CNV probes to support robust copy number calling. At 37.3 Mb, it is a comprehensive panel that allows you to accurately identify and profile novel and known variants associated with cancer.

Reliable sequencing for primary, metastatic or liquid biopsies
We know that you may want to use the same exome product on multiple sample types – especially when, for example, carrying out tumor/normal comparisons. The Cell3™ Target: Tumor Exome kit has been developed for and validated on a broad range of sample types, including FFPE, fresh frozen, genomic and cfDNA.
Low input FFPE and cfDNA sequencing is achievable from as little as 1 ng of genomic or cfDNA. With optimized protocols for FFPE our tumor exome kit unlocks the door for many oncology applications.
Detect low frequency variants in primary tumors
The Cell3™ Target library preparation behind our tumor exome enrichment incorporates error suppression technology. This includes unique molecular indexes (UMIs) and unique dual indexes (UDIs), to remove both PCR and sequencing errors and index hopping events.
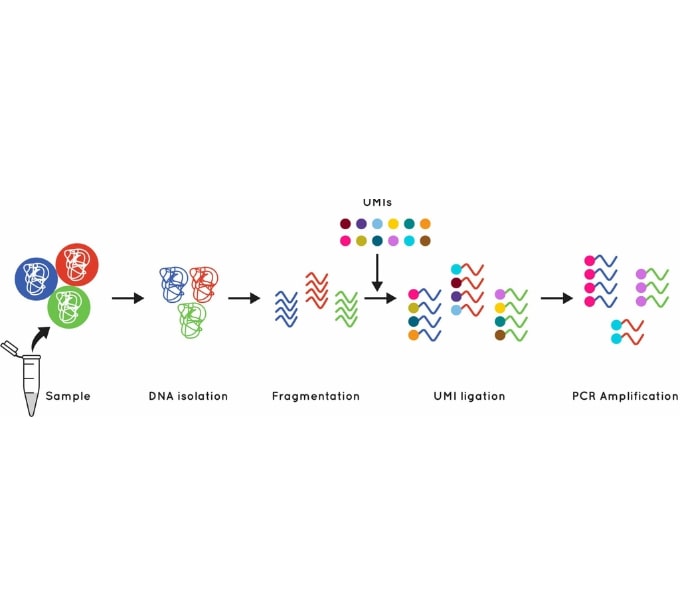
Figure 1. Using UMI’s to identify and quantify individual DNA molecules during library preparation increases sensitivity.
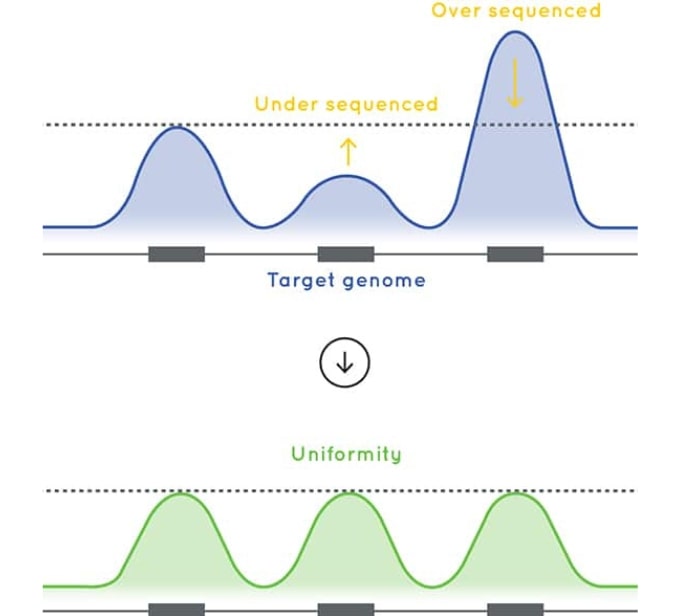
Optimized sequencing performance
The baits used in Cell3™ Target: Tumor Exome are designed to deliver excellent uniformity of coverage. By improving uniformity of coverage and reducing the number of low coverage exons, our tumor exome enrichment optimizes sequencing efficiency and sample capacity per sequencing run.
Customisable content for cancer profiling
Designed to be flexible, our Cell3™ Target: Tumor Exome allows you to add extra content specific to your project. Whether this is additional content or increased coverage of existing content, our Panel Design Tool makes this a simple and easy process to implement. And our rapid production turnaround means you will receive a fully NGS-validated custom exome panel within weeks.
- Additional content with high enrichment uniformity
- Increased coverage of specific genes covered by Cell3™ Target: Whole Exome
- Optimization of spike-in ratio.
Start your custom design now using the Panel Design Tool
See our customer publications
Cell3™ Target panels are available with one of two versions of our library preparation kits:
- Fragmentation: for use with DNA (genomic, FF, FFPE)
- Non-fragmentation: for use with cell-free DNA
| Product | Catalog No. |
| Cell3™ Target: Tumor Exome, Frag 16 samples | NGS_C3T_TEX_FR_16 |
| Cell3™ Target: Tumor Exome Frag 96 Samples | NGS_C3T_TEX_FR_96_A/B/C/D* |
| Cell3™ Target: Tumor Exome, Non Frag 16 samples | NGS_C3T_TEX_NF_16 |
| Cell3™ Target: Tumor Exome, Non Frag 96 samples | NGS_C3T_TEX_NF_96_A/B/C/D* |
* To provide flexibility in multiplexing samples, our 96-sample kits offer a choice in adapter plate:
A = Adapter plate with indexes 1-96
B = Adapter plate with indexes 97-192
C = Adapter plate with indexes 193-288
D = Adapter plate with indexes 289-384