Key features
- Scalable extraction to suit your laboratory needs, with a range of input volumes from 0.5-10 ml
- Fully automated or manual protocol compatible with most open-source liquid handlers
- High binding capacity beads that maximise cfDNA recovery
- Elution volumes as low as 30 µl ensuring cfDNA is concentrated enough for downstream applications without the need for DNA vacuum concentration
- Reversible nucleic acid-binding properties of paramagnetic particles along with an optimised binding system that targets recovery of small cfDNA fragments and minimises gDNA contamination
cfDNA extraction from plasma or serum
The utility of cfDNA in research has increased dramatically since its discovery.
Extracting cfDNA from biological samples can be challenging due to its low concentration and small fragment size. It is critical to minimise genomic DNA (gDNA) contamination from white blood cells, to enable reliable downstream analysis. Therefore, a cfDNA isolation method that is highly efficient at recovering smaller fragment sizes is highly desirable.
Nonacus have developed a flexible and scalable bead-based cfDNA extraction kit that is able to quickly and efficiently isolate high quality cfDNA from up to 10 ml plasma or serum samples.
Flexible cfDNA purification
The Bead Xtract cfDNA kit is simple and easy to use and is completely scalable to fit with your sample requirements. A flexible input volume of 0.5-10 ml plasma or serum avoids the requirement for multiple cfDNA extractions, saving time and laboratory resources.
The Bead Xtract cfDNA protocol offers ultimate user flexibility, as it can be run manually or as a fully automated process and is highly compatible with a range of open-source liquid handlers.
Maximise cfDNA yield
The high binding capacity of the paramagnetic particles used in the Nonacus Bead Xtract cfDNA kit maximises cfDNA yield from plasma samples. It also reduces the volume of particles required to isolate the cfDNA so that an elution volume as low as 30 µl can be achieved even with very large volumes of plasma.
High performance cfDNA extraction
The Bead Xtract cfDNA kit and protocol are optimised for recovering smaller DNA fragments ensuring that cfDNA yield is maximised and gDNA contamination is minimised.
We compared the DNA extraction yield (ng) of the Bead Xtract cfDNA extraction kit to a range of spin-column and bead-based cfDNA extraction kits currently available on the market. DNA extraction was performed manually using 1 ml plasma replicates following the individual company instructions and DNA quantification showed that the Bead Xtract cfDNA kit outperformed the other extraction methods (Figure 1).
Figure 2 shows typical nucleosomal patterning indicative of successful cfDNA isolation and minimal gDNA contamination. The high quality cfDNA extracted is suitable for downstream applications such as quantitative PCR (qPCR), droplet digital PCR (ddPCR) and Next Generation Sequencing (NGS)
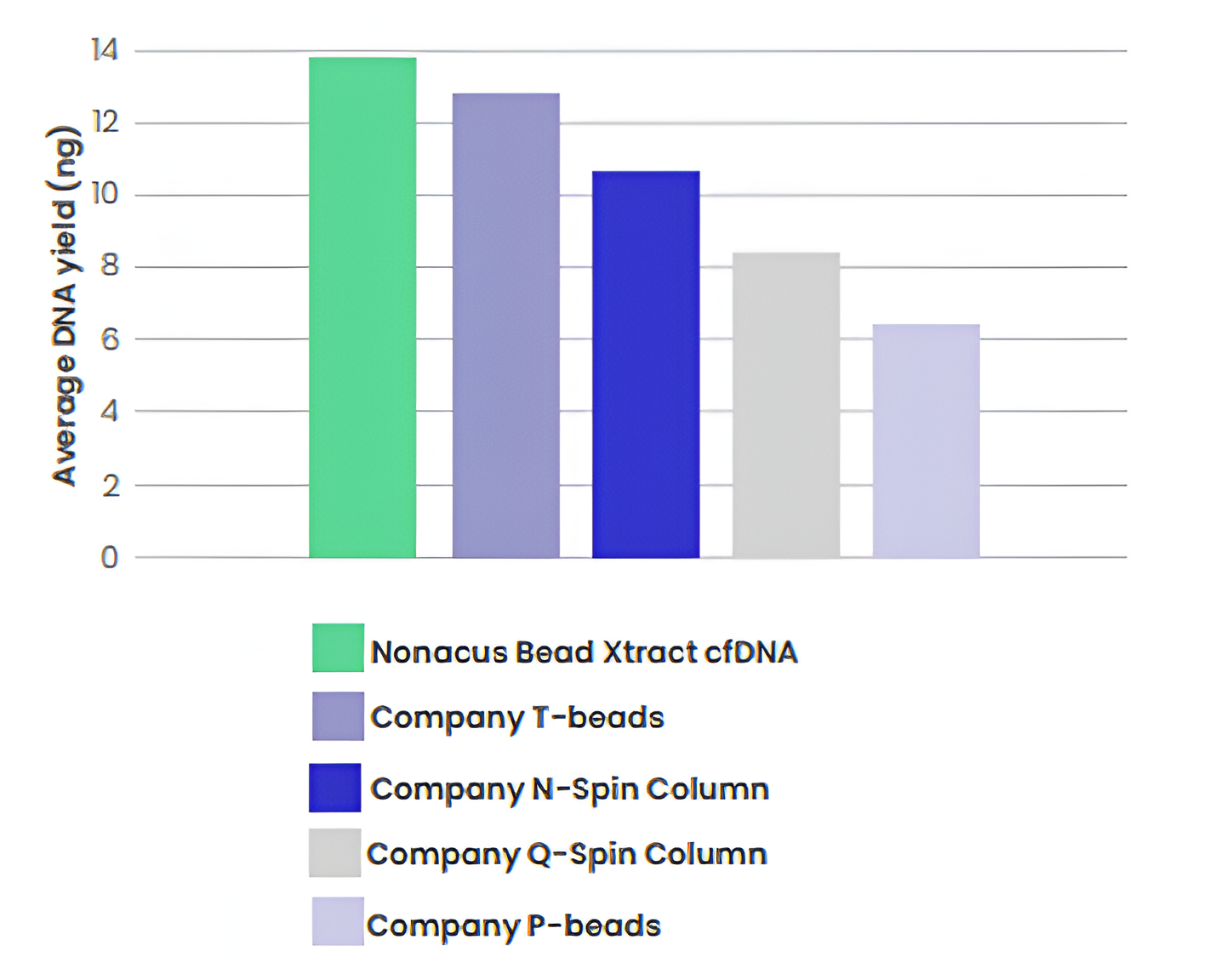
Figure 1: Average DNA yield (ng) extracted from 1 ml plasma replicates using a range of different bead-based and spin-column based extraction kits. cfDNA was extracted following the manufacturer's instructions and eluted into 50 µl of elution buffer. DNA concentration was determined using high sensitivity reagents on the Qubit 3.0 (Invitrogen).
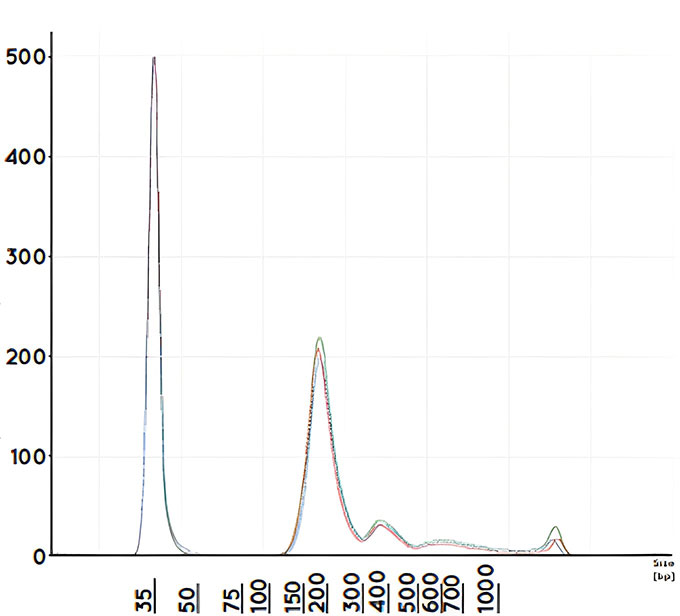
Figure 2: Automated cfDNA extraction using the Bead Xtract cfDNA kit shows expected nucleosomal patterning typical of cfDNA fragment distribution and low gDNA contamination. 4 ml replicates performed on the Hamilton ML STAR and eluted in 50 µl of buffer. Purified DNA was analyzed on the Agilent 4200 Tapestation
Streamlined workflow
The properties of the paramagnetic particles in the Bead Xtract cfDNA kit ensure fast magnetic separation even when using large volumes of plasma.
The elimination of funnels and vacuum steps streamlines the process so that manual extraction takes less than 70 minutes with a hands-on time of under 30 minutes.
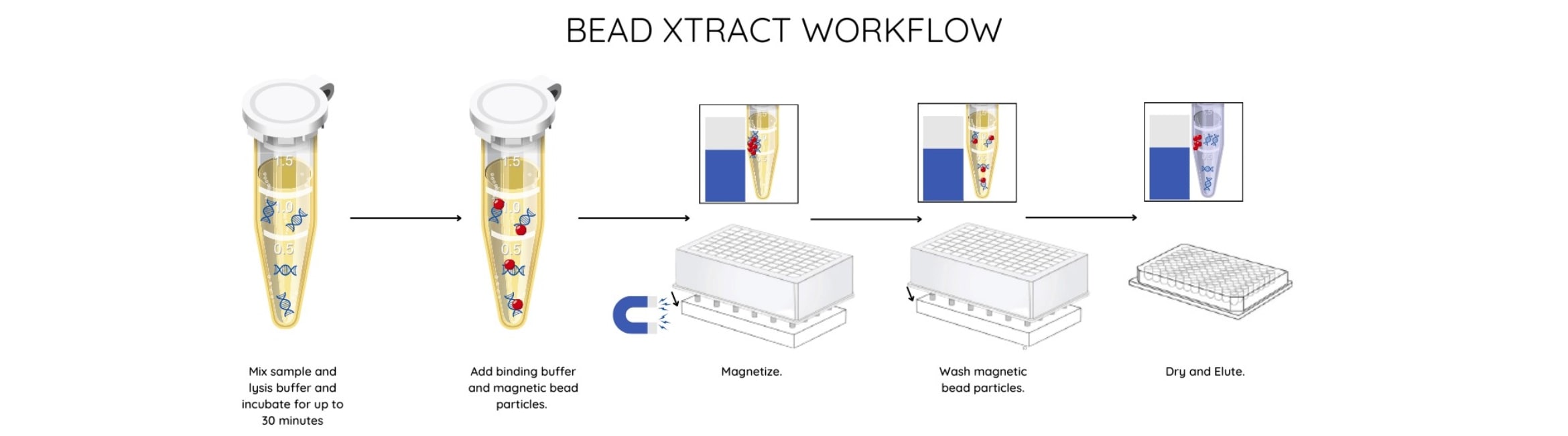
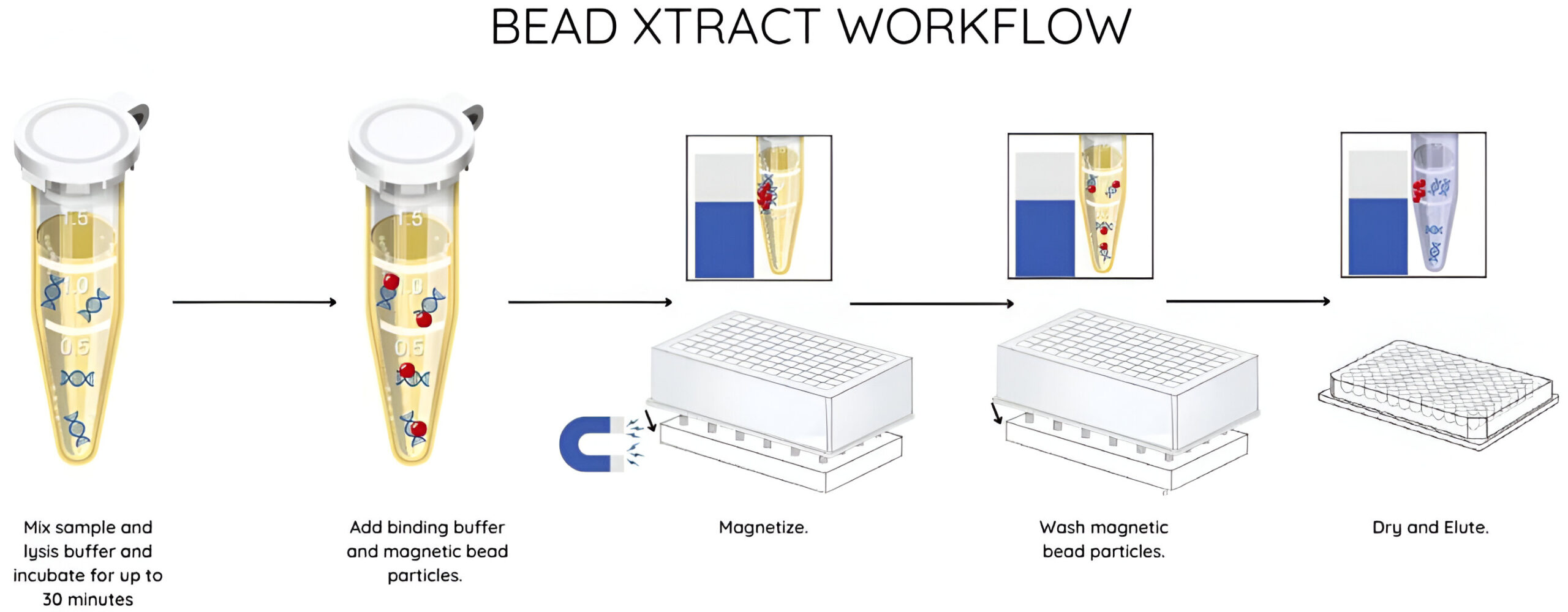
Automated cfDNA extraction
The Bead Xtract cfDNA protocol offers users flexibility to be run manually or as a fully automated process to suit laboratory and sample throughputs. Technical validation of automated isolation from lysis to extraction has been performed on the Hamilton ML STAR but protocols and scripts are available for a range of open-ended liquid handlers upon request.
Please contact our technical support team for more information.
Fully automated cfDNA extraction requires just 30 minutes set-up time and has been shown to generate highly concordant results with manual processing.

For Research Use Only. Not for use in diagnostic procedures.
