Unique molecular identifiers: the key to unmasking real low frequency variants
Written by Victoria Simms, January 28, 2022. Reviewed by Celina Whalley. April 26, 2024

How molecular barcodes help remove the PCR and sequencing errors that can mask important low-frequency variations in the analysis of cell-free DNA.
Next Generation Sequencing (NGS) technologies are powerful tools that have accelerated the rate of life science research for the last decade. This is especially true in the areas of oncology and prenatal genetics where the analysis of cell-free DNA (cfDNA), DNA that is released into the blood by either a tumor (ctDNA) or a fetus (cffDNA), has opened up the possibility of non-invasive patient testing and monitoring.
However, due to the low abundance of cfDNA, its analysis is challenging as it requires highly sensitive and accurate techniques to call variants with high confidence. Targeted sequencing, where the sequencing focuses on a number of genes or regions of interest, is one of those techniques. Cost effective and without the data burden of whole-genome sequencing (WGS) or whole-exome sequencing (WES), targeted sequencing allows researchers to achieve very high depths of coverage. This provides increased sensitivity and accuracy; but it is not without its flaws.1 Sequencing low-abundance DNA at high depth also increases the level of sequencing artefacts, creating background noise and potentially masking low-frequency variants. In light of this, it is essential when you are working with cfDNA that extra care is taken to ensure that errors are suppressed and only high-quality variant calls are retained.
The use of unique molecular identifiers (UMIs) - also known as molecular barcodes (MBCs) - is a recognised method for error suppression. Easily introduced as a simple step within your sequencing library preparation, UMIs can significantly reduce the background noise created by PCR and sequencing errors and enable mutation calling of variant allele frequencies (VAFs) down to 0.1%. This is especially important when deploying the ultra-deep sequencing necessary for the analysis of cfDNA.
Errors from PCR amplification and sequencing
Most typical library preparation protocols rely on PCR amplification of your starting DNA to increase the number of molecules to an adequate amount for sequencing. As PCR amplification is not a perfect process, errors will be introduced into the DNA copies, potentially introducing artefacts or 'false mutations' that could be confused in downstream analysis for real low frequency variants. The more cycles of amplification required prior to sequencing, the more likely PCR errors will be introduced. When working with low-abundance samples like ctDNA and cffDNA, or low-quality DNA (e.g. FFPE samples), the number of amplification cycles required may be 2-3 times that of a normal library preparation, making these sample types more prone to PCR-induced error.
Sequencing is not a perfect process either and the native error rate varies between sequencing technologies from 0.1-15%. Consequently, the more times you sequence a DNA fragment (eg. the ultra-deep targeted sequencing of cfDNA), or the more error prone the sequencing technology (eg. long-read sequencing generated by single molecule sequencing technologies), the more likely a sequencing error will be introduced.
PCR duplicates
When sequencing multiple PCR copies originating from the same DNA molecule, the resulting reads are referred to as PCR duplicates.2 Increasing the number of PCR cycles during your library preparation firstly increases the number of PCR duplicates for any given DNA fragment and secondly increases the possibility of introducing error into those duplicates. Sequencing experiments from samples such as ctDNA or cffDNA can give PCR duplicates at a rate of 50-60%.1 However, this can reach up to 90% when sequencing at depths of 20,000x or more (unpublished in-house data).
False positives from PCR duplicates and sequencing errors
PCR and sequencing-induced errors derived from PCR duplicates commonly occur with a low concentration or quality of starting material, as in the case for ctDNA, cffDNA and FFPE DNA, as these sample types require both increased PCR cycles for library amplification and ultra-deep sequencing. These 'errors' must be corrected for in your downstream analysis to prevent misinterpreting the data. Including them will not only lead to an over estimation of coverage in the duplicated region but also, and more importantly, incorrect allele frequency estimations and the creation of false positives where the error is interpreted as a minor allele.
Identifying PCR duplicates and removing them from a sequence analysis helps you to distinguish true sequence variants from potential false-positive results.
PCR duplicate removal
PCR duplicate removal is a common step in bioinformatic pipelines (e.g. SAMTools; Picard) where reads that align to the exact same mapping start point are removed.2 These bioinformatic methods for PCR duplicate removal are, however, simplistic, and whilst they flag duplicate reads that could arise through biases in the PCR process, they also flag genuine counts from different input molecules. They are useful to use when sequencing complexity is low, as when performing targeted sequencing of ctDNA, is it important to use UMIs.
So, what are UMIs and why are they useful?
UMI is a molecular tag consisting of a short known DNA sequence that is used to identify and quantify unique DNA molecules. These short molecular tags are ligated to the end of your DNA fragments during library preparation, before PCR amplification. This gives each initial input DNA molecule its own unique tag (Fig.1).
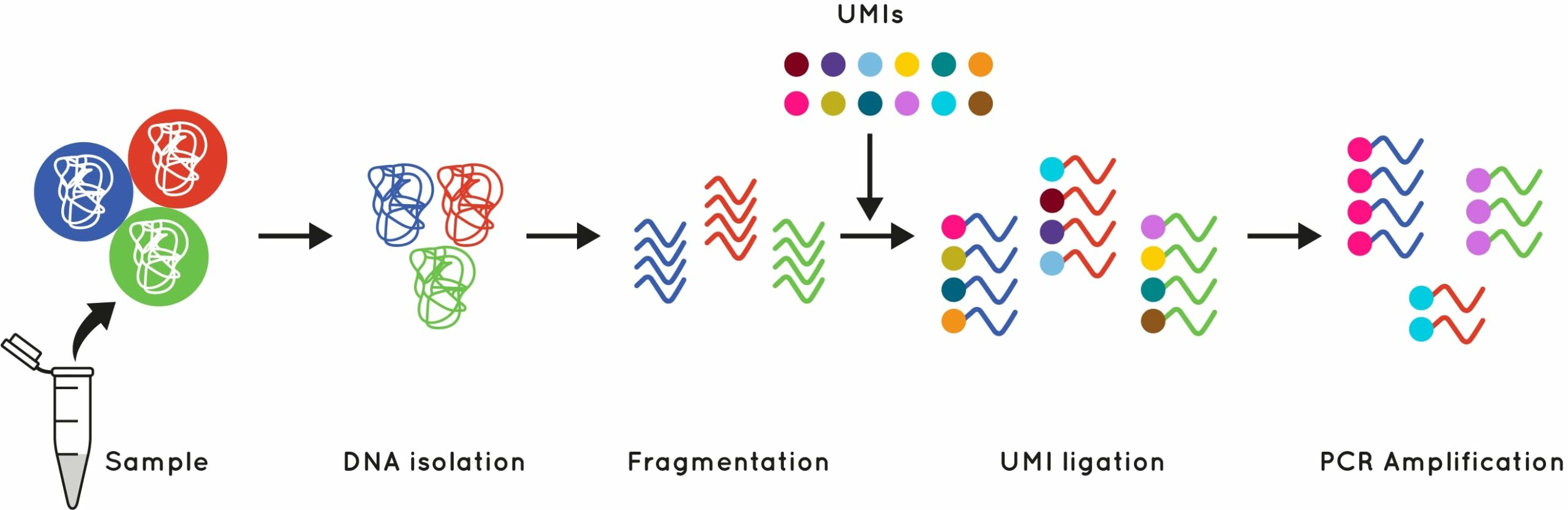
Figure 1: What is a UMI?
Sequencing reads containing the same UMI, that map to the identical genomic location, are assumed to originate from the same DNA molecule and are considered to be PCR duplicates. These reads can be grouped into a ‘consensus family’ (Figure 2.). If a variant occurs in all reads in the same family, then the consensus read sequence will include that variant. However, if a variant is only detected in a fraction of the reads in the family, it will be considered an error and disregarded.2
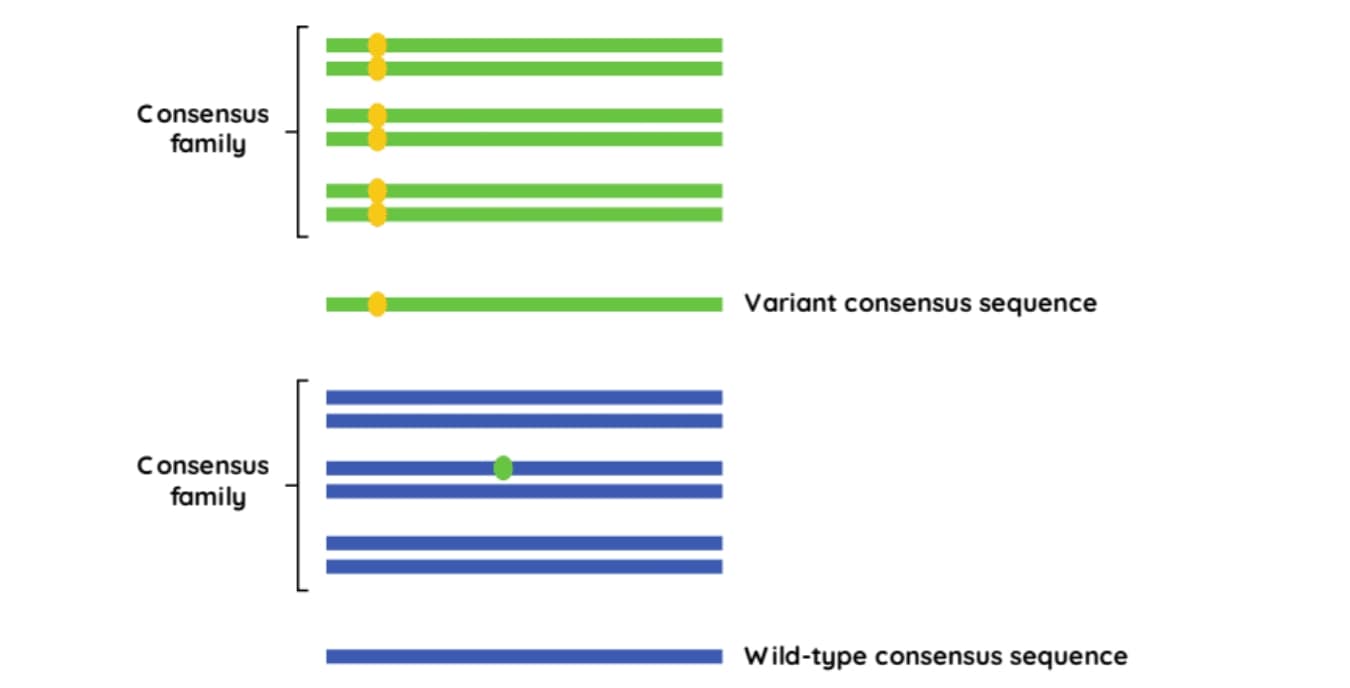
Figure 2: Using UMIs to create consensus families.3
UMIs ensure that only true duplicate reads are consolidated into consensus families. Unrelated reads with the same coordinates will have a different UMI and will be treated as unique (Figure 3).
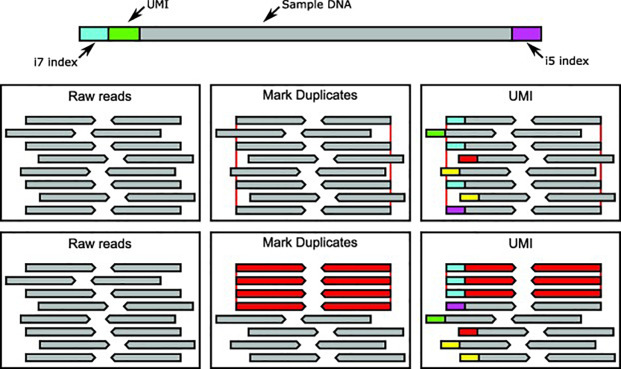
Figure 3: Methods of correcting for PCR duplicates in NGS.
The above diagram describes two approaches in dealing with duplicates in ultra-deep sequencing. In the top middle and right boxes duplicate reads are highlighted by the red lines at each end of the read. In the middle and right lower boxes, duplicate reads are red and unique reads are grey. Each UMI is indicated as the coloured block at the start of each read, with the UMI approach correctly discriminating the true duplicates. In this way, the UMI approach allows for ultra-sensitive variant detection in very deep sequencing applications.
UMI based de-duplication is able to leverage the high levels of PCR duplicates seen with ultra- deep targeted sequencing of cfDNA to create confidence in consensus reads and allows for ultra-sensitive variant detection in applications like cancer genome sequencing3 and foetal genetics.4
When and how to use UMIs
UMIs very efficiently and effectively remove the background noise created by PCR- and sequencing-associated errors that can mask true results when performing the ultra-deep targeted sequencing required for detecting low VAFs in low-abundance DNA.5,6
UMIs are appropriate for applications that produce high levels of duplicates. For, example, when sequencing low input cfDNA to detect very low frequency variants, ultra-deep sequencing is necessary (20,000 - 30,000 raw read depth). This results in high duplicates (~90%). When duplicates are removed using the UMI approach this will result in a consensus read depth of 2,000 - 3,000x. Cell3 Target's built in UMIs enables a single workflow for all sample types and tests allowing confident and sensitive calling of mutations down to 0.1% VAF and enables generation of sequencing libraries from as little as 1 ng of cfDNA input. This allows you to choose to use them or not without any penalties in your resulting data. However, UMIs are not necessary when sequencing at lower depths, eg. sequencing gDNA from whole blood or from tissue samples (FFPE or FF). This is because the proportion of duplicates is much lower (approx. 4% when sequencing gDNA at 100x depth for the ExomeCG).
UMI ligation is a simple, affordable step that is easily added to your normal library preparation protocol. During data analysis, reads with the same UMI are grouped into families and condensed into single consensus reads with all the duplicates reads removed. This filtering is simple to incorporate into any downstream bioinformatics pipelines, for example; Gencore, Connor and CGAT.
Cell3™ Target technology
The advancement of non-invasive techniques for oncology and prenatal genetics requires high-quality variant calls from low-abundance sample types, like ctDNA and cffDNA, to inform clinical decisions. The use of targeted sequencing is a cost-effective way of doing this but the need for a greater number of PCR cycles in library preparation, and the depth to which a sample must be sequenced to achieve meaningful results, means an increase in PCR duplicates, error and false positives. Error suppression technology, like UMIs or MBCs, is therefore critical to ensuring that the sequence data you obtain is sufficiently sensitive and accurate for the technique to be of value to patients and healthcare professionals alike. That is why Nonacus have incorporated UMIs into their Cell3 Target library preparation kits - so you don't need to worry about PCR or sequencing errors creating false-positive results and you can be confident in your sequencing data. Cell3 Target's built-in UMIs enable a single workflow for all sample types and tests, and you can choose to use them or not without any penalties in your resulting data.*
*There are applications where UMIs are less useful, for example, in sequencing genomic DNA for the detection of constitutional mutations or in sequencing DNA derived from FFPE where sequencing depth requirements are lower. However, you can choose not to use them in the downstream analysis without any penalty to your sequencing data.
Abbreviations
ct: circulating tumor
cf: cell-free
cff: cell-free fetal
FFPE: Formalin Fixed Paraffin Embedded
MBC: Molecular barcode
NGS: Next generation sequencing
WES: Whole exome sequencing
WGS: Whole genome sequencing
UMI: Unique molecular identifier
VAF: Variant allele frequency
References
- Bewicke-Copley F, Kumar EA, Palladino G, Korfi K, Wang J. Applications and analysis of targeted genomic sequencing in cancer studies. Computational and structural biotechnology journal. 2019;17:1348-59.
- Ebbert MT, Wadsworth ME, Staley LA, Hoyt KL, Pickett B, Miller J, et al. Alzheimer’s Disease Neuroimaging Initiative, Kauwe JS, Ridge PG. Evaluating the necessity of PCR duplicate removal from next-generation sequencing data and a comparison of approaches. BMC bioinformatics. 2016;17:491-500.
- Mansukhani S, Barber LJ, Kleftogiannis D, Moorcraft SY, Davidson M, Woolston A, et al. Ultra-sensitive mutation detection and genome-wide DNA copy number reconstruction by error-corrected circulating tumor DNA sequencing. Clinical chemistry. 2018;64(11):1626-35.
- van Campen J, Silcock L, Yau S, Daniel Y, Ahn JW, Ogilvie C, et al. A novel non‐invasive prenatal sickle cell disease test for all at‐risk pregnancies. British journal of haematology. 2020;190(1):119-24.
- Smith T, Heger A, Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome research. 2017;27(3):491-9.
- Chin RI, Chen K, Usmani A, Chua C, Harris PK, Binkley MS, et al. Detection of solid tumor molecular residual disease (MRD) using circulating tumor DNA (ctDNA). Molecular diagnosis & therapy. 2019;23(3):311-31.