4 key factors influencing NGS custom panel design
Written by Celina Whalley on December 8, 2023 | Reviewed by Victoria Simms on April 24, 2024
Question:
Why is good NGS panel design important for targeted sequencing?
Answer:
Effective probe-set design ensures comprehensive target coverage and heightened sensitivity. A poor design can lead to missed opportunities in detecting critical genetic variants.
So, what makes for good panel design? This is a short question with a complex answer, as good design ultimately comes down to your unique research question, but this highlights why custom panel design is so powerful.
Targeted sequencing enables you to focus efforts on specific regions of interest (ROI), enabling rare genomic variant detection with high specificity and sensitivity. Added benefits to this are cost streamlining and time saving.
Building a custom NGS panel can be challenging. Still, excellent online panel design tools are available to you with flexible input options for gene lists and/or specific genomic coordinates, but you still need a starting point.
Alongside colleague input, a good starting point for sourcing advice are academic articles, as your genes of interest may have already been successfully designed and the data published. Bear in mind how fast literature can change so a reliable panel design tool is invaluable to make probe-set adjustments in response to any revisions.
Here we have reviewed four key factors to consider when designing your panel, so that you can make the most of these online tools.
Key panel design considerations:
1- Choosing the right target regions
2- Sequencing coverage vs depth
3- Reducing experimental noise
4- Validating your design
1- Choosing the right target regions
Online panel building tools will require you to input your ROIs, this can be in the format of gene lists or genomic co-ordinates. These need to relate directly to your research objectives – for example, are you looking for specific variants or structural changes within whole genes or hotspots?
Aspects to consider include:
- Relevance: Ensure that the selected ROIs are biologically relevant to your research question
- Coverage: Evaluate the coverage of genes within ROIs to avoid missing critical information
- Challenging regions: Highly homologous regions, regions of high and low GC content, homopolymers, and repeats of any size cannot be sequenced with high confidence or quality.1 This can lead to inadequate coverage, and the gathering of non-specific information, so assess the relative importance of these regions
- Exonic vs. Intronic: Decide whether you want to sequence protein coding regions only or include intronic regions for more comprehensive analysis
- Known vs. novel variants: are you interested in targeting known disease, or drug, associated variants or discovering novel ones?
- Striking the right balance with panel size v cost: Larger panels include more content with a higher number of probes leading to greater sequencing capacity and cost. Too few probes would be cheaper to sequence, but you could possibly miss crucial variants reducing the sensitivity of your panel
- Genome selection: selecting the latest version will provide the most updated genome scaffold to design probes from. For example, the most frequently used human genome builds are - GRCh37 and GRCh38. So which one should you use?
- GRCh38 - The latest human reference genome is the Genome Research Consortium human build 38 (GRCh38) also known as Human Genome build 38 (Hg38).2 This build was published in 2013 and contains corrected sequencing artifacts, fewer gaps, and more alternate loci compared with the previous GRCh37 (b37/Hg19) assembly3
- GRCh37 – This older version human reference genome was released in 2009. Many research projects will have used this as their reference genome so it should be considered used if your research project is a continuation of their work
2- Sequencing coverage vs depth
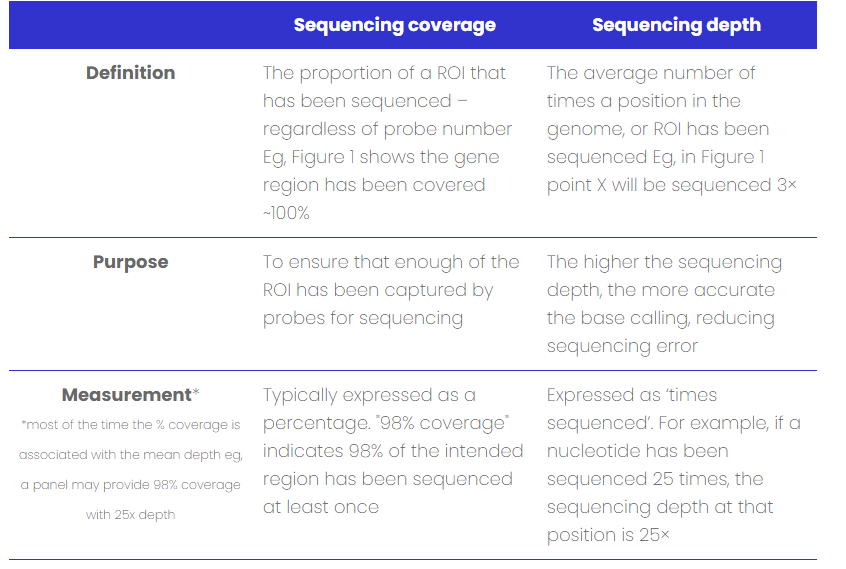
What is coverage? Clinicians and genomic researchers need to consider sequencing depth and coverage when planning projects and analysing genomic data. The two factors are related but are often used interchangeably despite their distinctions,4 see Figure 1, and Table 1. Read on to find out the difference between the two.

Figure 1: A schematic overview defining sequencing depth and coverage across a single ROI.
Table 1: Differences between sequencing coverage and depth.
How do you take coverage into account when designing your panel?
- Tiling options
- Off-target effects
Tiling options
What is tiling? In the context of custom panel design "tiling" refers to the strategic arrangement of probes covering your ROIs. The goal is to minimize gaps and overlaps, ensuring that every part of the region of interest is effectively captured.
Panel design tools will offer choice on the level of tiling and Figure 2 shows how probe numbers increase alongside tiling levels ie, if you have chosen 1x tiling and then choose 2x, your probe numbers will double across the ROI.
The higher the number of probes that cover a genomic position, the greater the robustness in variant calling as the opportunity for its capture is increased. However, this is a balancing act, as an increased number of probes will increase associated sequencing costs - which is why validation of your panel on your real-world samples prior to large scale projects is so important.
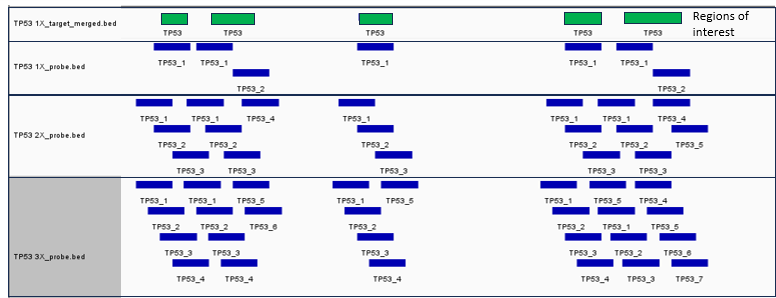
Figure 2: Adapted from IGV software to show how probe numbers alter between 1x, 2x and 3x tiling designs (green bars are the ROIs and the blue bars are the probes).
Additional flexibility: check to see if the panel design tool has options to gap fill, or automatically add pre-validated probes to your genes/regions of interest.
For example, when using the Nonacus Panel Design Tool the option for ‘gap filling’ automatically includes pre-validated probes designed within tricky masked regions of the genome.
Automatic addition of pre-validated probes within unmasked regions can also be achieved by choosing the ‘exome’ option. Both options build strength into the design as the probes are tried and tested within the commercially available Cell3 Target: Whole Exome Enrichment panel.
Off-target effects
Off-target effects can be defined as the capturing and sequencing of unintended regions. These need to be minimized as the impact can lead to wasted sequencing capacity and increased un-necessary costs. To reduce off-target sequencing a well-designed panel should take into account:
- Stringent probe design strategies to minimize off-target capture, ensuring that sequencing efforts are focused on the intended regions only
- Masking repetitive regions that would result in probe failure, or mis-priming
- Algorithms built into the design tool to ensure uniformity of coverage preventing fluctuation in sequencing depth between targeted regions. See Figure 3.
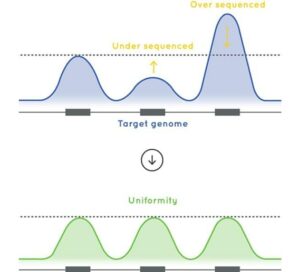
Figure 3: A schematic overview showing the benefit uniformity of coverage preventing sequencing bias between targeted regions.
Learn more: Nonacus Custom Panel Design Tool
Sequencing depth
How deep should you sequence? There are no fixed recommendations for sequencing depth, this is unique to your panel design. Most users will determine best practice defined within the scientific community, and literature related directly to their area of interest.
Depth needs to be high enough to detect all variants (SNVs, indels, structural variants and CNVs) without the introduction of sequencing errors, which can occur at too-low depths adding to any experimental noise factors, which then pass into bioinformatic pipelines, misdirecting study conclusions.4
3- Reducing experimental noise
What does ‘experimental noise’ mean? Experimental noise encompasses the impact of library assay kits, PCR duplicates and sequencing errors in NGS data, all of which will introduce anomalies influencing your final conclusions.
To minimize these risks, locate an online panel design tool from a company that provides a complete NGS solution from panel creation and manufacture, through to library preparation assays.
Why? At Nonacus, for example, the online probe design tool compliments the Cell3 Target library preparation assay, which handles experimental noise factors automatically by:
- Expanding the range of starting material; validated for all DNA types – gDNA, cell-free DNA and FFPE – down to 1 ng input
- Eliminates PCR and sequencing errors with unique molecular identifiers (UMIs). Please read our blog for further insight into the power of UMIs
- Prevents index-hopping during sequencing with dual indexing5
- Boosts conversion rates maximizing the amount of successfully ligated DNA suitable for sequencing
4- Validating your design
Before committing to sequencing, validate your panel's performance using control samples with known genotypes to identify and rectify issues with primer design, off-target effects, or suboptimal coverage.
With Nonacus panel designs, an NGS quality check is included, technically validating each probe, but biological validation can only be achieved with your real-world patient samples, so trial your design on a small sample sub-set prior to any larger study.
Conclusion
Empower your research with NGS custom design. A well-designed custom panel is cost-effective, maximizes coverage, minimizes off-target effects, reduces noise, and offers flexibility and scalability.
We are driven at Nonacus to help overcome challenges surrounding NGS panel design and our Panel Design Tool, coupled with Cell3 Target library preparation assays, aims to enhance discoveries in cancer diagnostics, longitudinal disease monitoring, and personalized medicine.
Table 2 below provides an overview of the benefits of our panel design tool, so contact the team for more information and to discuss your ideas. We look forward to hearing from you.
Table 2: How Nonacus supports custom NGS design.

Customer publications using the Nonacus custom panel design tool:
- Evidence for Early Detection Of Metastatic Variants and Tumour Evolution in ctDNA of Patients with Oesophageal Adenocarcinoma
- NGS Custom Panel Implementation in Patients with Non-Syndromic Autism Spectrum Disorders in the Clinical Routine of a Tertiary Hospital
- Targeted deep sequencing of urothelial bladder cancers and associated urinary DNA: a 23-gene panel with utility for non-invasive diagnosis and risk stratification
- 2024. Mutation detection in saliva from oral cancer patients.
References
1. Bean LJH, Funke B, Carlston CM, et al. Diagnostic gene sequencing panels: from design to report—a technical standard of the American College of Medical Genetics and Genomics (ACMG). Genetics in Medicine. 2020;22(3):453-461. doi:10.1038/S41436-019-0666-Z
2. Homo sapiens genome assembly GRCh38.p14 - NCBI - NLM. Accessed October 31, 2023.
3. Homo sapiens genome assembly GRCh37 - NCBI - NLM. Accessed October 31, 2023.
4. Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. Sequencing depth and coverage: key considerations in genomic analyses. nature.com D Sims, I Sudbery, NE Ilott, A Heger, CP Ponting Nature Reviews Genetics, 2014 Published online 2014. doi:10.1038/nrg3642
5. Index Hopping | Intro & how to minimize it. Accessed November 20, 2023.