The art of maximizing cell-free DNA yield from plasma
July 14, 2021. Reviewed by Celina Whalley, April 26, 2024

Cell-free DNA testing: What you need to consider before you start
A successful next generation sequencing assay using cell free DNA (cfDNA) relies heavily on the pre-analytical steps; sample collection, storage and cfDNA extraction. The pre-analytical products developed at Nonacus have been designed to maximize both cfDNA quality and yield.
In fact pre-analytical elements are considered so important that a list of Recommended Preanalytic Data Elements, including 11 required preanalytic attributes that are essential for studies, is included in the BloodPAC Data Commons (BPDC); an open, publicly accessible data commons for the global liquid biopsy community. BloodPAC (Blood Profiling Atlas in Cancer) was launched in 2016 to accelerate the development, validation and clinical use of liquid biopsy assays.
Using cell-free DNA for non-invasive genetic testing or liquid biopsy assays
The introduction of non-invasive, or minimally invasive methods, using cfDNA and next generation sequencing (NGS) for molecular diagnostics is now more widespread in research and clinical medicine. This is especially true in the fields of oncology and prenatal genetics where the analysis of cfDNA, which is released into the blood by either a tumor or a fetus, can provide valuable information for healthcare professionals.
A simple blood test (liquid biopsy) can help determine a patient's suitability for a specific drug, monitor their response to treatment or provide a prenatal genetic diagnosis, all bringing real improvements to patient care. However, due to the low abundance of cfDNA, a successful test using this relies heavily on the pre-analytical steps - the collection, storage and processing of samples.
The importance of correct procedures in blood sample collection and storage
Whilst taking a blood sample is a simple, non-invasive procedure, any contamination of the cfDNA fraction with intracellular genomic DNA (gDNA), shed into the plasma by the degradation of nucleated cells will affect the reliability and validity of your data. This is particularly important for both oncology and prenatal applications, where the accurate detection of very low variant allele frequencies (VAF) is critical and the presence of any gDNA could mask true results.
Importantly, Lippi et al. report that approaching 60-70% of pre-analytical mistakes are derived from incorrect procedures in the collection, storage and processing of specimens.1
A recent study by Lampignano et al. aimed to establish standardized pre-analytical workflows for quality assurance and reported on the impact of pre-analytics (cfDNA extraction and quantification) in downstream mutation detection.2 They determined that the two critical aspects contributing to the success of a genetic test using cfDNA are the quality and quantity of the extracted material.
Here we offer tips and guidance on the three main steps in the pre-analytical procedure for cfDNA extraction prior to genetic analysis:
1) Blood collection and storage
2) Plasma collection and storage
3) cfDNA extraction, storage and quality control (QC) measurements
1. Blood collection and storage
There are two types of blood collection tubes suitable for plasma extraction, those containing additives that stabilize blood cells and those without. Whilst tubes without stabilization additives can be used, these must be spun within 24 hours of blood draw and kept on ice until first centrifugation. This protects against the lysis of nucleated cells (white blood cells) and the subsequent release of gDNA into the plasma sample.
As this is not always practical, especially with regard to sample transport, tubes containing stabilisation additives, like the Cell3™Preserver tubes, can be used instead. The stabilizer in these tubes preserves the nucleated cells (white blood cells) in an intact state and helps in the anti-coagulation of blood cells preventing the release of intracellular gDNA into plasma and any change in the cell-free DNA fraction during transport. These allow for the blood sample to be stored for up to 21 days and at an ambient temperature before processing.
Figure 1 demonstrates the stability of cfDNA concentration measurements up to 21 days post blood-draw using Cell3™ Preserver blood collection tubes.
Following the first centrifugation, which is performed to separate the plasma from the buffy coat and red blood cells, the plasma is centrifuged for a second time at 10,000 g or higher to remove cell debris and any remaining intact cells in the sample.
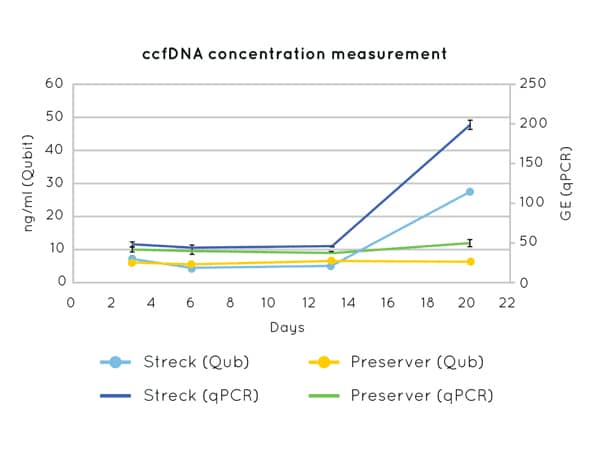
Figure 1: DNA concentration measurements by Qubit (left axis) and qPCR (CCR5 amplification) (right axis) on cfDNA extracted from plasma collected in Nonacus Cell3™ Preserver and Streck tubes and isolated at 3, 6, 13 and 20 days post blood draw. Streck tubes show an apparent increase in cfDNA concentration from day 13 due to gDNA released by the lysis of contaminating white blood cells.
As there is only a low concentration of cfDNA in plasma, it is important as well as minimising gDNA contamination, to start with adequate volumes of whole blood. In fact the predominant challenge for molecular diagnostics in both oncology and prenatal genetics is that an even smaller and variable fraction of cfDNA - potentially as little as 1% - is derived from either the tumor (circulating tumor DNA (ctDNA)) or the fetus (circulating free fetal DNA (cffDNA)).
To enable detection of low frequency variants by NGS in either of these examples, a sample volume of 8 ml of whole blood is recommended as a starting amount. Approximately half of that volume, 4 ml, should be recovered as plasma, so you may expect, in a healthy individual, to recover 1-5 ng of cfDNA per ml of plasma.
2. Plasma collection and storage
If you obtain 4 ml plasma per 8 ml tube of whole blood, then aim to use the whole sample for extraction. It is recommended to store plasma at -20˚C for short term storage (weeks) or -80˚C for long term storage (months to years) in aliquots to avoid freeze-thaw cycles and to extract cfDNA from plasma only when you are ready to start your NGS library preparation or cfDNA test.
3. cfDNA extraction, storage and QC measurements
There are many commercially available kits, both bead and column based, for the extraction of cfDNA from plasma. Page et al made comparisons between four different cfDNA extraction methods and detected wide variation between these in terms of yield.3
High cfDNA yield does not necessarily mean a better extraction and could indicate the presence of gDNA or carrier RNA which is used in some commercial extraction kits to enhance the recovery of DNA. Appropriate QCs are advised and running an aliquot of the extracted nucleic acid on a TapeStation ScreenTape will indicate the level of gDNA/carrier RNA contamination in the sample. Accurate quantification of cfDNA is important for downstream applications including NGS library preparation.
Extracted cfDNA can be stored at -20˚C however freeze-thaw cycles should be avoided, and samples should be accurately quantified once thawed prior to starting your NGS library preparation or cfDNA test.
A successful cell-free DNA assay
Working with cfDNA for either oncology or prenatal applications requires the extremely sensitive and accurate measurement of genomic alterations. The two critical aspects contributing to the success of a genetic test using cfDNA are the quality and quantity of the extracted material as the presence of any gDNA could mask true results. Owing to the relative scarcity of cfDNA, the importance of the pre-analytical steps of sample collection, storage and processing cannot be underestimated.1
More information on pre-analytical products from Nonacus; Cell3 Preserver, Bead Xtract cfDNA, Cell3™Xtract is available here:
References
- Lippi G, Chance JJ, Church S, Dazzi P, Fontana R, Giavarina D, et al. Preanalytical quality improvement: from dream to reality. Clinical Chemistry and Laboratory Medicine. 2011;49(7):1113-26.
- Lampignano R, Neumann MH, Weber S, Kloten V, Herdean A, Voss T, et al. Multicenter evaluation of circulating cell-free DNA extraction and downstream analyses for the development of standardized (pre) analytical work flows. Clinical Chemistry. 2020;66(1):149-60.
- Page K, Guttery DS, Zahra N, Primrose L, Elshaw SR, Pringle JH, et al. Influence of plasma processing on recovery and analysis of circulating nucleic acids. PloS One. 2013;8(10):e77963.