The importance of using an optimized pre-analytical workflow for liquid biopsy analysis
Written by Victoria Simms, February 14, 2023. Reviewed by Celina Whalley.

Do you use cell-free DNA as a biomarker for clinical diagnostics or medical research?
Find out how using a workflow specifically designed for liquid biopsies can help improve the quantity and quality of your cell-free DNA samples.
Liquid biopsies are increasingly used to guide clinical decisions in the cancer healthcare field. Although circulating tumor cells and exosomes can be studied as liquid biopsy analytes, it is often circulating nucleic acid known as cell-free DNA (cfDNA) and tumor derived cfDNA (ctDNA) that are used as biomarkers for disease.
Through genetic and epigenetic evaluation, ctDNA can reveal valuable insights into tumor stage and location, as well as indicating a patient's suitability for specific treatment options. The majority of cfDNA is released via cell death into the bloodstream, therefore to monitor tumor status or detect minimal residual disease (MRD) using cfDNA, regular liquid biopsy samples are often taken from patients.
In healthy individuals, cfDNA concentrations range from 0-100 ng/ml1, whereas in cancer patients this concentration can be considerably higher, correlating with tumor stage and size.2 Although the total cfDNA concentration is higher in these individuals, the ctDNA fraction can constitute as little as 0.01%. This large amount of background cfDNA means that it is often challenging to detect low frequency variants from ctDNA.
Owing to the relative scarcity of ctDNA and the challenge of detecting it against a background of normal cfDNA, a pre-analytical workflow that uses specially designed products to protect and stabilize cfDNA cannot be underestimated.
It is reported that 60-70% of pre-analytical mistakes occur from incorrect procedures within the stages of sample collection, storage and processing.3 In this blog, we cover the key considerations at each stage of the pre-analytical workflow (Figure 1), which will help you to maximize your cfDNA yield and enable sensitive detection of low frequency variants.
The pre-analytical workflow consists of:
- Blood collection and storage
- Plasma preparation and storage
- Cell-free DNA extraction
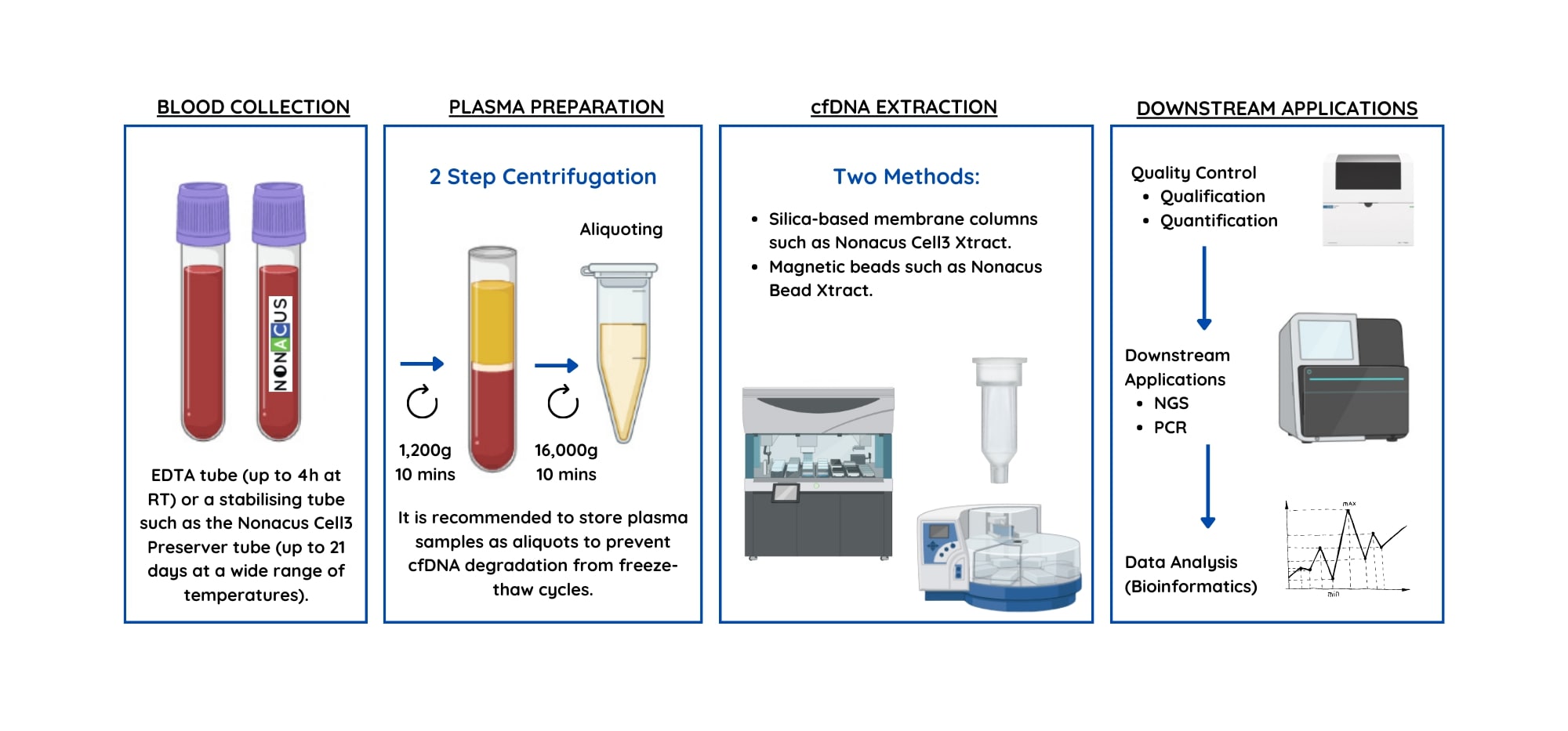
Figure 1: The pre-analytical process included the steps of blood sample collection, plasma preparation and cell-free DNA extraction.
What are the key considerations at each stage of the liquid biopsy workflow?
1. Blood sample collection
There are several factors during the liquid biopsy collection stage which have a direct impact on the quality and quantity of the final cfDNA sample. As reported by the ESMO Precision Medicine Working Group, these considerations include:
- Timing of blood sampling
- Volume of blood sample collected
- Type of collection tube used
- Sample storage conditions
Careful consideration should be given to the timing of sample collection, since patient specific factors such as: the type and timing of treatment, surgery and inflammation levels will all impact the concentration of cfDNA in the blood. To avoid the likelihood of false-negative results during genetic variant analysis, ESMO recommend that blood sampling is avoided whilst individuals undergo active therapy and for MRD detection, samples should be collected at least 1 week after surgery.4
The blood sample volume should be planned alongside the timing of sample collection, as cfDNA concentration directly correlates with the volume of plasma that the cfDNA will be extracted from.
For cfDNA recovery from plasma samples, a blood sample is normally collected via venipuncture into blood collection tubes (BCTs). The type of BCT used, will be dependent on the liquid biopsy analyte being studied, as well as the time between collection and processing, sample transportation and temperature conditions.
Using standard EDTA tubes as BCTs prevents coagulation of the blood but requires the sample to be kept at temperatures of ~4oC and processed within four hours. These strict requirements often limit their use to internal studies.
Blood collection tubes developed specifically for cfDNA analysis, like the Nonacus Cell3™ Preserver tubes (Figure 2), contain stabilization additives as well as EDTA. This prevents the lysis of hematopoietic (white blood) cells - a critical aspect in reducing gDNA contamination in the final cfDNA sample.
By preventing white blood cell lysis, BCTs like the Nonacus Cell3 Preserver tubes allow whole blood samples to be stored for up to 21 days at a wide range of temperatures (4-37oC) (Figure 3). This minimizes the adverse effects of time, storage, and transport conditions on sample integrity and cfDNA extraction yield allowing laboratories to send and receive blood samples from customers or partner labs across the world.
Figure 2: Nonacus Cell3 Preserver Blood Collection Tube.
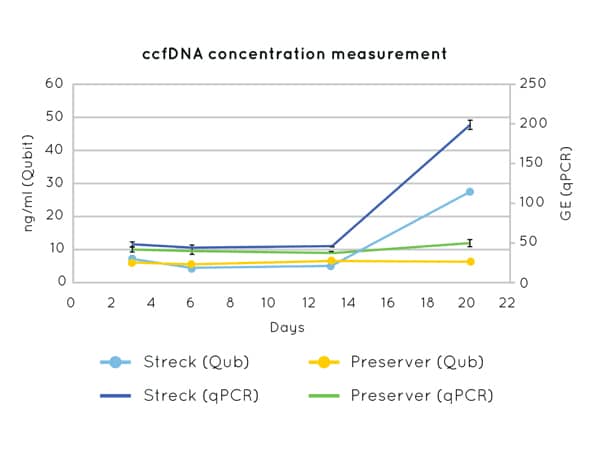
Figure 3: DNA concentration measurements by Qubit (left axis) and qPCR (CCR5 amplification) (right axis) on cfDNA extracted from plasma collected in Nonacus Cell3 Preserver and Streck tubes and isolated at 3, 6, 13 and 20 days post blood draw. Streck tubes show an apparent increase in cfDNA concentration from day 13 due to gDNA released by the lysis of contaminating white blood cells.
2. Plasma preparation
After liquid biopsy collection, the plasma fraction must be isolated from the whole blood sample. This can be done by centrifugation and the protocol at this stage will influence cfDNA yield and the likelihood of contaminating factors like intact cells and cellular debris, being in the final cfDNA sample which is used for NGS library preparation. It is advised to use a two-step centrifugation method to obtain a plasma fraction that is free from cellular material.
Since cfDNA exists in low abundance, adequate volumes of plasma must be obtained, which is mainly influenced by the initial blood sample volume. As a guide, for variant detection analysis using NGS it is recommended that at least 8 mls whole blood is collected, as this will equate to ~4 mls of plasma in the first stage of the plasma isolation process.
The following centrifugation protocol is recommended by Nonacus:
- Step one: centrifuge the whole blood sample at 2000g for 10 minutes - to separate the plasma fraction from the red blood cells and the buffy coat containing white blood cells (shown in Figure 4).
- Step two: centrifuge the plasma fraction at >10,000g for 10 minutes - to remove cellular debris from the plasma fraction.
If cfDNA extraction does not immediately proceed plasma preparation, the plasma sample should be stored at -20oC for short term storage (weeks) or at -80oC for longer term storage (months-years). To prevent cfDNA degradation, care should be taken to minimize temperature fluctuations and freeze-thaw cycles.
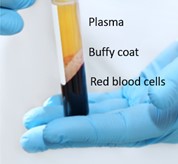
Figure 4: During the first stage of plasma isolation, the plasma fraction will be separated from the buffy coat containing the white blood cells and the red blood cells.
3. Cell-free DNA extraction
A key element to the success of downstream applications of PCR and NGS is the quantity of the extracted cfDNA. It is for this reason that the type of extraction method, along with the volume of plasma used for extraction needs careful consideration.
It is advised to extract cfDNA only when you are ready to perform the subsequent downstream applications to prevent any deterioration of sample and unnecessary freeze-thaw cycles. The process of extracting cfDNA involves removing proteins and other biological components from the plasma sample, and can be done via two different methods:
- Bead-based extraction
- Spin-column extraction
The type of cfDNA extraction method you choose will depend on your specific sample and laboratory requirements.
Bead-based cfDNA extraction methods like the Nonacus Bead Xtract cfDNA kit use magnetic beads to target the recovery of small DNA fragments such as cfDNA (Figure 5). This is important as it minimizes gDNA contamination in the final cfDNA sample, which if present could mask true results. Many bead-based extraction kits can be run on liquid handlers as well as manually. This ability to automate processing offers user flexibility, as well as saving precious processing time.
Spin-column based cfDNA extraction methods use a centrifugation protocol alongside silica-based membrane columns to isolate cfDNA. Spin-based extraction methods like the Nonacus Cell3 Xtract kit enable cfDNA extraction from a range of biological sample types including plasma, serum, cerebral spinal fluid, saliva, and amniotic fluid.
Regardless of the cfDNA extraction method, for cases such as early cancer detection, a high volume of plasma in some cases as much as 10 ml may be required to maximize cfDNA yield. Both cfDNA extraction kits provided by Nonacus (Bead Xtract and Cell3 Xtract) allow for a flexible input volume of up to 10 ml and use low elution volumes to ensure cfDNA is concentrated enough without the need for DNA vacuum concentration steps.
Extracted cfDNA should be stored at -20˚C and freeze-thaw cycles should be avoided to prevent cfDNA deterioration.
Accurate quantification of cfDNA is always needed ahead of downstream applications like NGS library preparation. The amount of cfDNA recovered will be sample and patient dependent, but as a guide it is expected that ~1-5 ng cfDNA can be recovered from ~1 ml of plasma (based on Nonacus Cell3 Xtract kit).
It is important to note that a high DNA yield post-extraction could indicate the presence of gDNA contamination or carrier RNA, which is sometimes used in some commercial extraction kits to enhance DNA recovery. Running a small amount of the extracted sample on a TapeStation to visualize the DNA profile could determine if this is the case. Within the trace, cfDNA extraction will be indicated by a peak at ~166 bp.5 and ctDNA may show up at a slightly lower size of ~145 bp.6
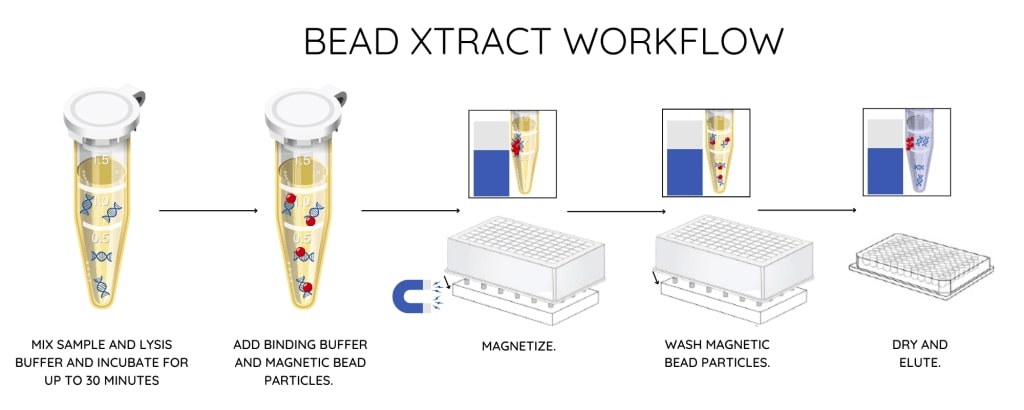
Figure 5: Bead-based cell-free DNA extraction protocol from the Nonacus Bead Xtract kit.
Molecular profiling of cancer can be achieved by analyzing ctDNA from liquid biopsies. In clinical practice, these results help to guide patient treatment decisions and ultimately lead to improved patient outcomes. Due to the low concentration of ctDNA within clinical samples and the requirement for ultrasensitive and accurate measurement of genomic alterations, the importance of using specifically designed liquid biopsy products cannot be underestimated. These products must ensure that the final cfDNA sample is both high in quality and quantity, to allow for accurate detection of low frequency variants.
For more information about the pre-analytical products offered by Nonacus, visit our pre-analytical products page which includes information on our Cell3 Preserver tubes, Bead Xtract cfDNA and Cell3 Xtract kits.
References
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nature Reviews Cancer. 2011;11(6):426-37.
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early-and late-stage human malignancies. Science translational medicine. 2014;6(224):1-25.
- Lippi G, Chance JJ, Church S, Dazzi P, Fontana R, Giavarina D, et al. Preanalytical quality improvement: from dream to reality. Clinical chemistry and laboratory medicine. 2011;149(7):1113-26.
- Pascual J, Attard G, Bidard FC, Curigliano G, de Mattos-Arruda L, et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: a report from the ESMO Precision Medicine Working Group. Annals of Oncology. 2022;33(8):750-68.
- Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164(1):57-68.
- Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Science translational medicine. 2018;10(466):1-28.